Calculates confidence intervals for one or more parameters in a fitted
model object. A function that returns the log-likelihood must be supplied,
either directly via the argument loglik or using a logLikFn S3
generic.
Usage
profileCI(
object,
loglik,
...,
parm = "all",
level = 0.95,
profile = TRUE,
mult = 32,
faster = TRUE,
epsilon = -1,
flat = 1e-06,
lb,
ub,
optim_args = list()
)Arguments
- object
A fitted model object. This object must have a
coefS3 method that returns a named vector of parameter values. Iffaster = TRUEorprofile = FALSEthen it must also have avcovS3 method that returns a matrix withdimnamesthat match those in the vector returned by thecoefmethod. If necessary, these may be created using.S3method(). For example, ifobjectis a list inheriting from class"foo", with coefficients inobject$coefficientsand variance-covariance matrix inobject$vcov, then use.S3method("coef", "foo", function(x) x$coefficients)and.S3method("vcov", "foo", function(x) x$vcov).- loglik
A named function that returns the log-likelihood based on input parameter values and data. The first argument must be the vector of model parameters. If the likelihood is zero for any observation in the data then the function should return
-Inf.Alternatively,
loglikdoes not need to be supplied if alogLikFnS3 method has been created forobject. TheprofileCIpackage provideslogLikFn.glm, which is used in an example in Examples.- ...
Further arguments to be passed to
loglik.- parm
A vector specifying the parameters for which confidence intervals are calculated, either a vector of numbers or a vector of names. The default,
which = "all", produces confidence intervals for all the parameters.- level
The confidence level required. A numeric scalar in (0, 1).
- profile
A logical scalar. If
TRUEthen confidence intervals based on a profile log-likelihood are returned. IfFALSEthen intervals based on approximate large sample normal theory, which are symmetric about the MLE, are returned.- mult
A positive numeric scalar. Controls the increment by which the parameter of interest is increased/decreased when profiling above/below its MLE. The increment is
mult * se / 100whereseis the estimated standard error of the estimator of the parameter. Decreasingmultprofiles at more points but will be slower.- faster
A logical scalar. If
faster = TRUEthen the profiling of the log-likelihood in search of a lower (upper) confidence limit is started at the corresponding symmetric lower (upper) confidence limit. However,faster = FALSEis more robust, particularly if a symmetric limit is close to, or beyond, the edge of the parameter space.- epsilon
Only relevant if
profile = TRUE. A numeric vector of values that determine the accuracy of the confidence limits.epsilonis recycled to the length of the parameter vectorparm.If
epsilon[i] > 0then this value is passed as the argumentepsilonto theitp::itpfunction, which estimates the parameter values for which the profile log-likelihood for parameteridrops to the value that defines the confidence limits, once profiling has been successful in finding an interval within which this value lies.If
epsilon[i] < 0monotonic cubic spline interpolation (stats::splinefunwithmethod = "hyman")is used, which will tend to be faster.If
epsilon[i] = 0then linear interpolation is used, which will be faster still.
- flat
A positive numeric scalar used to avoid continuing a search for a confidence limits in cases where the profile log-likelihood becomes flat. If a reduction in profile log-likelihood is less than
flat * mult / 100then the search is stopped. The value of the returned confidence limit isInffor an upper limit and-Inffor a lower limit.- lb, ub
Optional numeric vectors of length
length(parm). If supplied,lb[i]and/orub[i]place respective lower and upper bounds on the interval over which profiling takes place for parameterparm[i]. If a bound is reached before a confidence limit is determined or before the profile log-likelihood is determined to have become flat, then the relevant limit is returned asNA. Elementwise,lbmust be smaller than, andublarger than,coef(object).- optim_args
A list of further arguments (other than
parandfn) to pass tostats::optim. For example,optim_args = list(method = "BFGS", control = list(trace = 1))changes the method used from"Nelder-Mead"to"BFGS"and setstraceto provide the lowest level of tracing information.
Value
An object of class c("profileCI", "matrix", "array"). A numeric
matrix with 2 columns giving the lower and upper confidence limits for
each parameter. These columns are labelled as (1-level)/2 and
1-(1-level)/2, expressed as a percentage, by default 2.5% and 97.5%.
The row names are the names of the parameters supplied in parm.
If profile = TRUE then the returned object has extra attributes crit,
level and for_plot. The latter is a named list of length equal to the
number of parameters. Each component is a 2-column numeric matrix. The
first column contains values of the parameter and the second column the
corresponding values of the profile log-likelihood. The profile
log-likelihood is equal to the attribute crit at the limits of the
confidence interval. The attribute level is the input argument level.
If faster = FALSE or epsilon > 0 then the attributes lower_pars and
upper_pars are lists that provide, for each profiling, the values of the
parameters for the last maximisation of the log-likelihood.
A matrix with columns giving the object
c("profileCI", "matrix", "array").
Details
The default, epsilon = -1, should work well enough in most
circumstances, but to achieve a specific accuracy set epsilon to be
a small positive value, for example, epsilon = 1e-4.
The defaults mult = 32 and faster = TRUE are designed to calculate
confidence intervals fairly quickly. If the object returned from
profileCI is plotted, using plot.profileCI, then we will not obtain
a smooth plot of a profile log-likelihood. Setting faster = FALSE and
reducing mult, perhaps to 8 or 16 should produce a smoother plot.
The arguments flat1, lb and ub are provided to prevent a call to
profileCI hanging in a search for a confidence limit that will never be
found.
Examples
## From example(glm)
counts <- c(18, 17, 15, 20, 10, 20, 25, 13, 12)
outcome <- gl(3, 1, 9)
treatment <- gl(3, 3)
glm.D93 <- glm(counts ~ outcome + treatment, family = poisson())
confint(glm.D93)
#> Waiting for profiling to be done...
#> 2.5 % 97.5 %
#> (Intercept) 2.6958215 3.36655581
#> outcome2 -0.8577018 -0.06255840
#> outcome3 -0.6753696 0.08244089
#> treatment2 -0.3932548 0.39325483
#> treatment3 -0.3932548 0.39325483
confint.default(glm.D93)
#> 2.5 % 97.5 %
#> (Intercept) 2.7095672 3.37947764
#> outcome2 -0.8505027 -0.05800787
#> outcome3 -0.6707552 0.08478093
#> treatment2 -0.3919928 0.39199279
#> treatment3 -0.3919928 0.39199279
# A Poisson log-linear GLM logLikFn.glm S3 method is provided in profileCI
# so we do not need to supply loglik explicitly
prof <- profileCI(glm.D93)
prof
#> 2.5% 97.5%
#> (Intercept) 2.6958271 3.36656379
#> outcome2 -0.8576884 -0.06255514
#> outcome3 -0.6753594 0.08244109
#> treatment2 -0.3932489 0.39324886
#> treatment3 -0.3932489 0.39324886
# Supplying a Poisson GLM log-likelihood explicitly
poisson_loglik <- function(pars) {
lambda <- exp(model.matrix(glm.D93) %*% pars)
loglik <- stats::dpois(x = glm.D93$y, lambda = lambda, log = TRUE)
return(sum(loglik))
}
# This will be a bit slower than profile.glm() because glm.fit() is fast
prof <- profileCI(glm.D93, loglik = poisson_loglik)
prof
#> 2.5% 97.5%
#> (Intercept) 2.6958271 3.36656379
#> outcome2 -0.8576884 -0.06255514
#> outcome3 -0.6753594 0.08244109
#> treatment2 -0.3932489 0.39324886
#> treatment3 -0.3932489 0.39324886
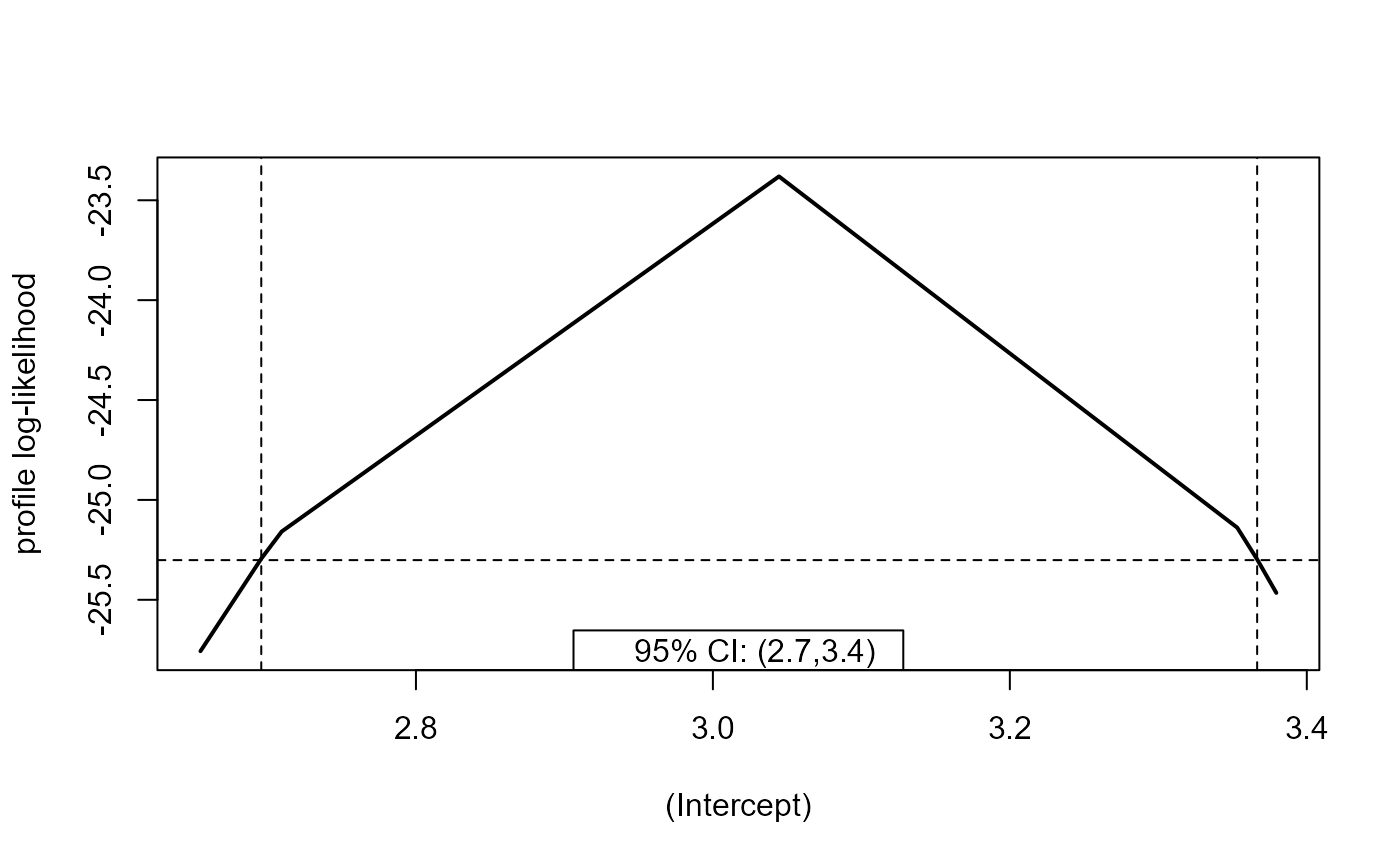
plot(prof, parm = 1)
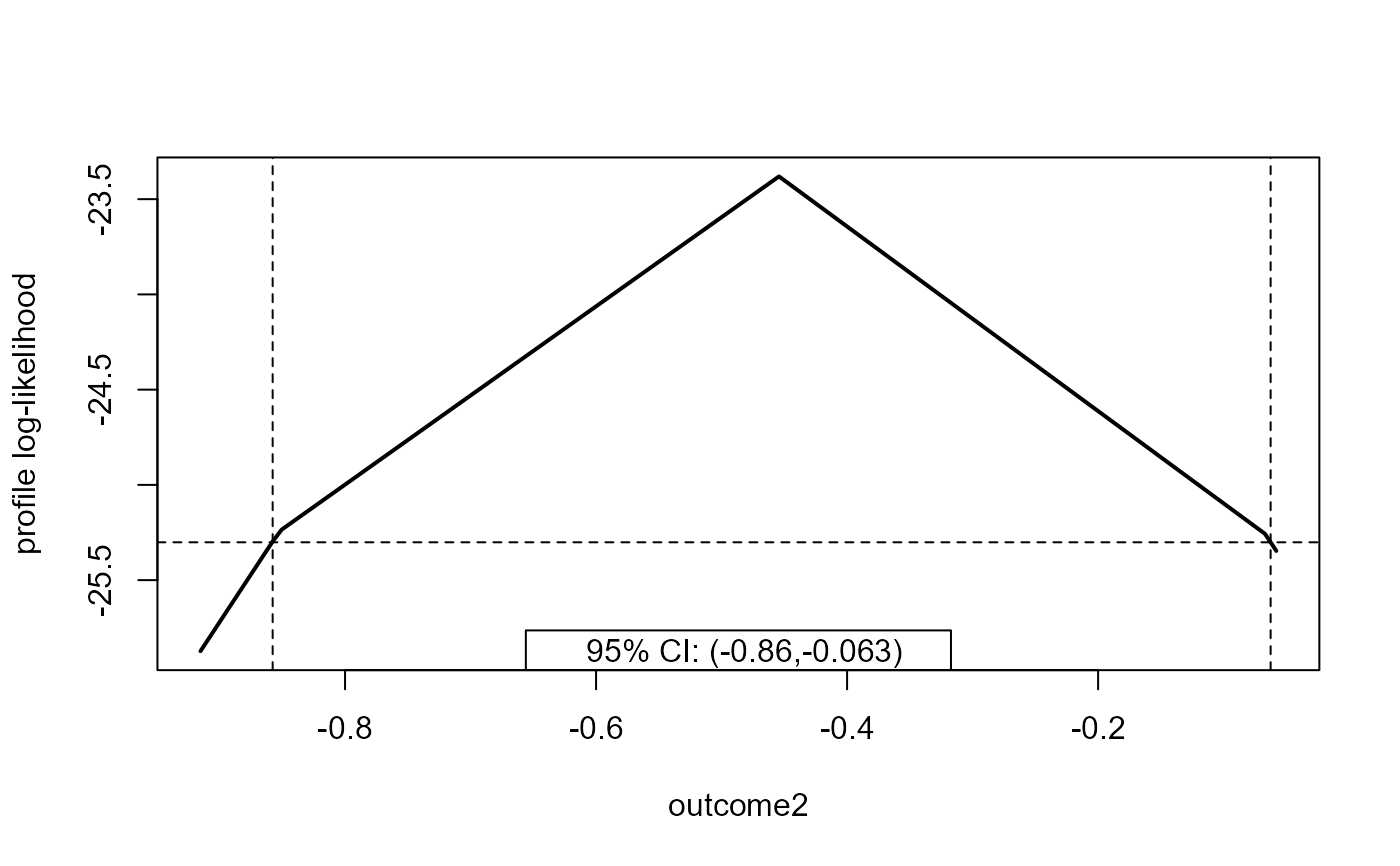
 plot(prof, parm = "outcome2")
plot(prof, parm = "outcome2")
 # Supplying a more general Poisson GLM log-likelihood
poisson_loglik_2 <- function(pars, glm_object) {
lambda <- exp(model.matrix(glm_object) %*% pars)
loglik <- stats::dpois(x = glm_object$y, lambda = lambda, log = TRUE)
return(sum(loglik))
}
prof <- profileCI(glm.D93, loglik = poisson_loglik_2, glm_object = glm.D93)
prof
#> 2.5% 97.5%
#> (Intercept) 2.6958271 3.36656379
#> outcome2 -0.8576884 -0.06255514
#> outcome3 -0.6753594 0.08244109
#> treatment2 -0.3932489 0.39324886
#> treatment3 -0.3932489 0.39324886
## Nonlinear least squares, from example(nls)
DNase1 <- subset(DNase, Run == 1)
fm1DNase1 <- nls(density ~ SSlogis(log(conc), Asym, xmid, scal), DNase1)
confint(fm1DNase1)
#> Waiting for profiling to be done...
#> 2.5% 97.5%
#> Asym 2.1935437 2.538804
#> xmid 1.3214540 1.678751
#> scal 0.9743072 1.114939
# profileCI() gives slightly different results because confint.nls() is
# not based on profiling the log-likelihood but rather changes in the RSS
prof <- profileCI(fm1DNase1)
#> Error in logLikFn.nls(object, pars = pars, ...): object 'DNase1' not found
prof
#> 2.5% 97.5%
#> (Intercept) 2.6958271 3.36656379
#> outcome2 -0.8576884 -0.06255514
#> outcome3 -0.6753594 0.08244109
#> treatment2 -0.3932489 0.39324886
#> treatment3 -0.3932489 0.39324886
# Supplying a more general Poisson GLM log-likelihood
poisson_loglik_2 <- function(pars, glm_object) {
lambda <- exp(model.matrix(glm_object) %*% pars)
loglik <- stats::dpois(x = glm_object$y, lambda = lambda, log = TRUE)
return(sum(loglik))
}
prof <- profileCI(glm.D93, loglik = poisson_loglik_2, glm_object = glm.D93)
prof
#> 2.5% 97.5%
#> (Intercept) 2.6958271 3.36656379
#> outcome2 -0.8576884 -0.06255514
#> outcome3 -0.6753594 0.08244109
#> treatment2 -0.3932489 0.39324886
#> treatment3 -0.3932489 0.39324886
## Nonlinear least squares, from example(nls)
DNase1 <- subset(DNase, Run == 1)
fm1DNase1 <- nls(density ~ SSlogis(log(conc), Asym, xmid, scal), DNase1)
confint(fm1DNase1)
#> Waiting for profiling to be done...
#> 2.5% 97.5%
#> Asym 2.1935437 2.538804
#> xmid 1.3214540 1.678751
#> scal 0.9743072 1.114939
# profileCI() gives slightly different results because confint.nls() is
# not based on profiling the log-likelihood but rather changes in the RSS
prof <- profileCI(fm1DNase1)
#> Error in logLikFn.nls(object, pars = pars, ...): object 'DNase1' not found
prof
#> 2.5% 97.5%
#> (Intercept) 2.6958271 3.36656379
#> outcome2 -0.8576884 -0.06255514
#> outcome3 -0.6753594 0.08244109
#> treatment2 -0.3932489 0.39324886
#> treatment3 -0.3932489 0.39324886