The bang package simulates from the posterior distributions
involved in certain Bayesian models. See the vignette Introducing bang: Bayesian Analysis, No
Gibbs for an introduction. In this vignette we consider the
hierarchical 1-way Analysis of variance (ANOVA) model:
where
,
so that
,
and
and all random variables are independent. Variability of the response
variable
about an overall level
is decomposed into contributions
from an explanatory factor indicating membership of group
and a random error term
.
This model has
parameters:
and
.
Equivalently, we could replace
by
.
The full posterior
can be factorized into products of lower-dimensional densities. See the
Appendix for details. If it is assumed that
and
are independent a priori and a normal prior is set for
then the only challenging part of sampling from
is to simulate from the two-dimensional density
.
Otherwise, we need to simulate from
.
The bang function hanova1 uses the rust
package (Northrop 2017) to simulate from
these densities. In sampling from the marginal posterior density
we use a parameterization designed to improve the efficiency of
sampling. In this case we work with
.
We illustrate the use of hanova1 using two example
datasets. Unless stated otherwise we use the default hyperprior
,
that is, a uniform prior for
for
(see Sections 5.7 and 11.6 of Gelman et al.
(2014)). A user-defined prior can be set using
set_user_prior.
Late 21st Century Global Temperature Projection Data
The data frame temp2 contains indices of global
temperature change from late 20th century (1970-1999) to late 21st
century (2069-2098) based on data produced by the Fifth Coupled Model
Intercomparison Project (CMIP5). The dataset is the union of four
subsets, each based on a different greenhouse gas emissions scenario
called a Representative Concentration Pathway (RCP). Here we analyse
only data for RCP2.6. Of course, inferences about the overall
temperature change parameter
are important, but it is also interesting to compare the magnitudes of
and
.
If, for example, if
is much greater than
then uncertainty about temperature projection associated with choice of
GCM is greater than that associated with the choice of simulation run
from a particular GCM. See Northrop and Chandler
(2014) for more information.
library(bang)
# Extract RCP2.6 data
RCP26_2 <- temp2[temp2$RCP == "rcp26", ]
There are 61 observations in total distributed rather unevenly across
the 38 GCMs. Only 28 of the GCMs have at least one run available for
RCP2.6.
# Number of observations
length(RCP26_2[, "index"])
#> [1] 61
# Numbers of runs for each GCM
table(RCP26_2[, "GCM"])
#>
#> ACCESS1-0 ACCESS1-3 bcc-csm1-1 bcc-csm1-1-m BNU-ESM
#> 0 0 1 1 1
#> CanESM2 CCSM4 CESM1-BGC CESM1-CAM5 CMCC-CM
#> 5 6 0 3 0
#> CMCC-CMS CNRM-CM5 CSIRO-Mk3-6-0 EC-EARTH FGOALS-g2
#> 0 1 10 2 1
#> FIO-ESM GFDL-CM3 GFDL-ESM2G GFDL-ESM2M GISS-E2-H
#> 3 1 1 1 1
#> GISS-E2-H-CC GISS-E2-R GISS-E2-R-CC HadGEM2-AO HadGEM2-CC
#> 0 1 0 1 0
#> HadGEM2-ES inmcm4 IPSL-CM5A-LR IPSL-CM5A-MR IPSL-CM5B-LR
#> 4 0 4 1 0
#> MIROC-ESM MIROC-ESM-CHEM MIROC5 MPI-ESM-LR MPI-ESM-MR
#> 1 1 3 3 1
#> MRI-CGCM3 NorESM1-M NorESM1-ME
#> 1 1 1
# Number of GCMs with at least one run
sum(table(RCP26_2[, "GCM"]) > 0)
#> [1] 28
We use hanova1 to sample from the posterior distribution
of the parameters based on the (default) improper uniform prior for
,
described in Section 11.6 of Gelman et al.
(2014). This prior fits in to the special case considered in the
Appendix, with an infinite prior variance
for
.
# The response is the index, the explanatory factor is the GCM
temp_res <- hanova1(resp = RCP26_2[, "index"], fac = RCP26_2[, "GCM"])
# Plots relating to the posterior sample of the variance parameters
plot(temp_res, params = "ru")
plot(temp_res, ru_scale = TRUE)
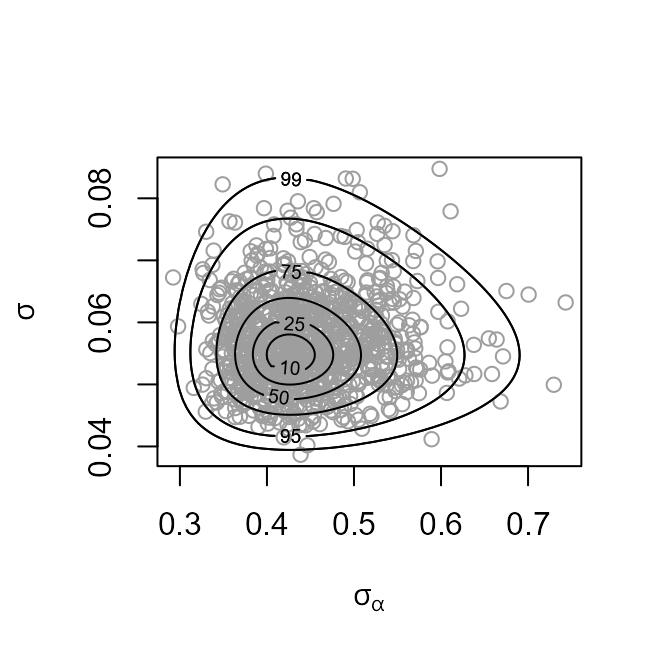
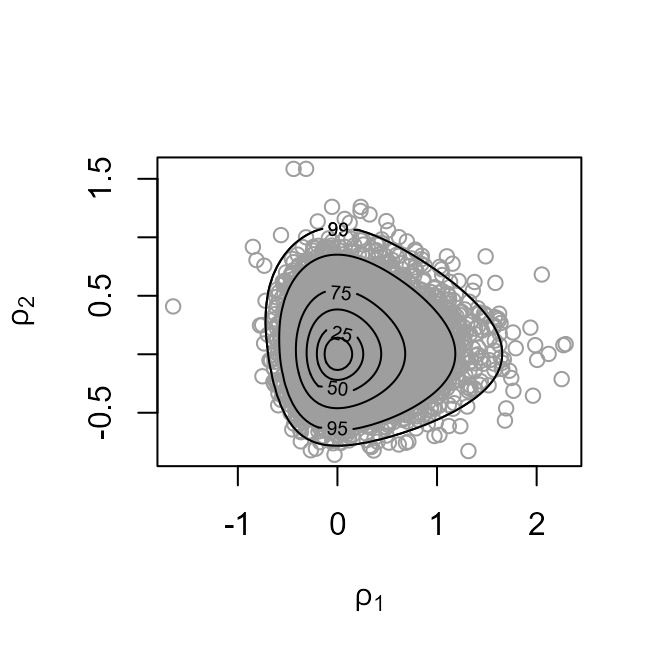
The plot on the left shows the values sampled from the posterior
distribution of
with superimposed density contours. We see that the posterior
distribution is located at values for which
is much greater than
,
by a factor of nearly 10. On the right is a similar plot displayed on
the scale used for sampling,
,
after relocation of the posterior mode to the origin and rotation and
scaling to near circularity of the density contours.
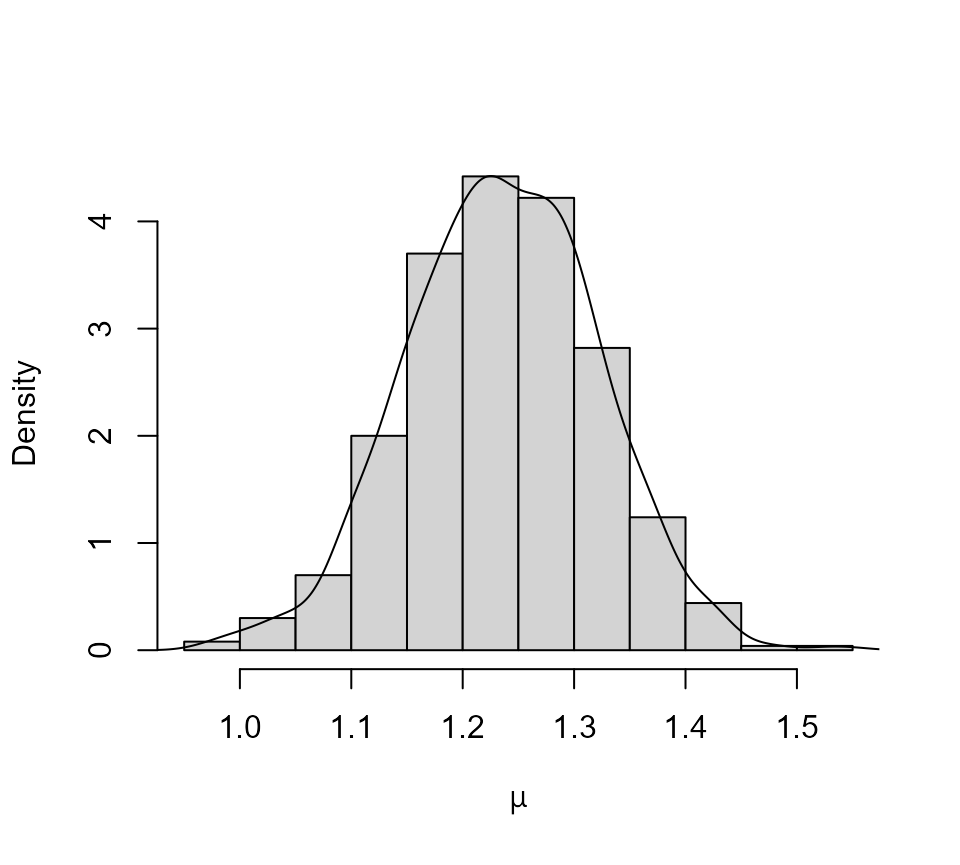
We summarize the marginal posterior distribution of
using a histogram with a superimposed kernel density estimate.
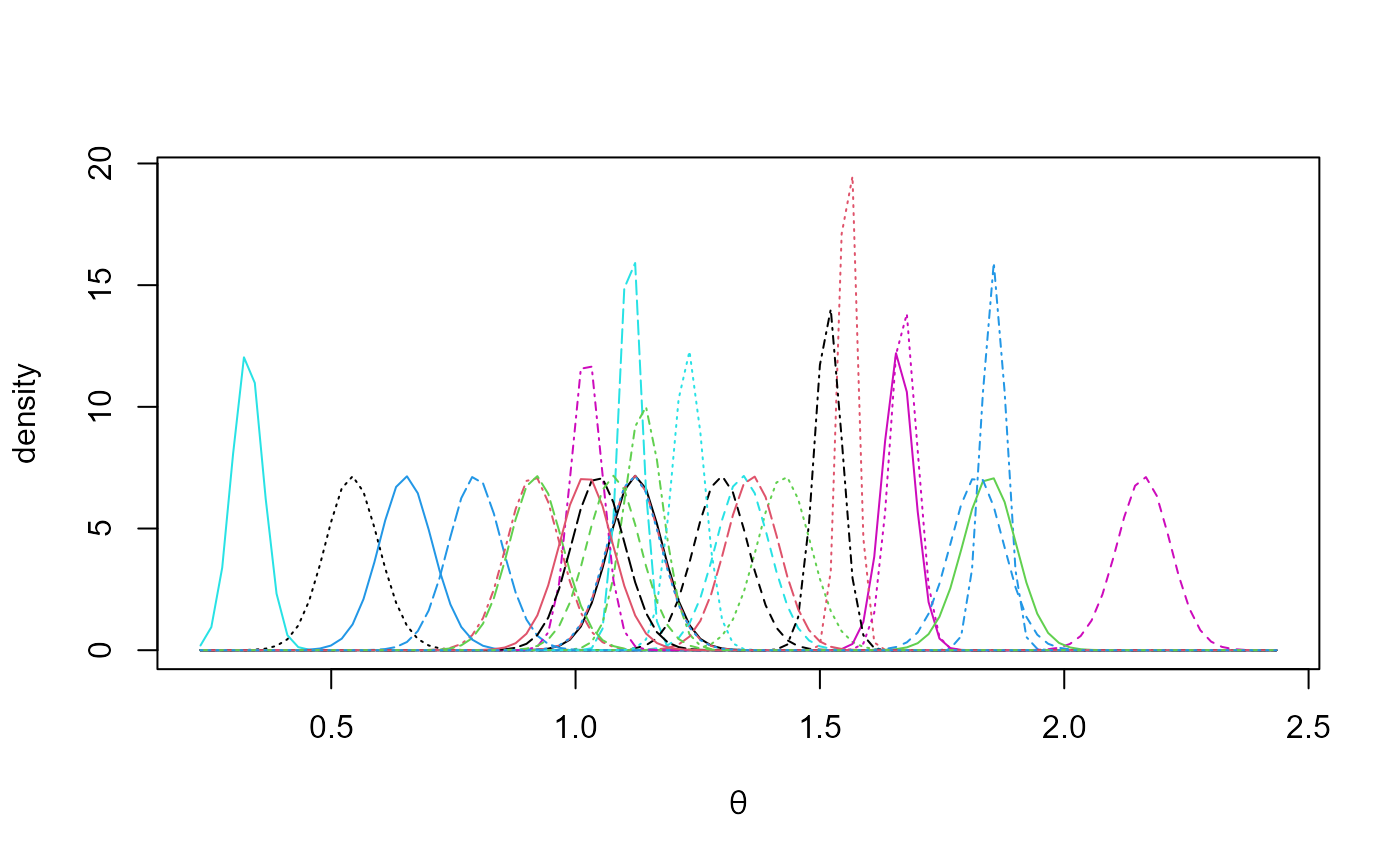
The following plot summarizes the estimated marginal posterior
densities of the mean index for each GCM.
plot(temp_res, params = "pop", which_pop = "all", one_plot = TRUE)
Coagulation time data
In the temperature projection example sampling was conducted on the
scale
and was unproblematic. It would also have been unproblematic had we
sampled on the original
scale. To show that there are examples where the latter is not the case
we consider a small dataset presented in Section 11.6 of Gelman et al. (2014). The response variable is
the coagulation time of blood drawn from 24 animals allocated to
different diets. The crucial aspect of this dataset is that the
explanatory factor (diet) has only 4 levels. This means that there is
little information about
in the data. Unless some prior information about
is provided the posterior distribution for
will tend to have a heavy upper tail (Gelman
2006).
The generalized ratio-of-uniforms method used by rust can
fail for heavy-tailed densities and this is indeed the case for these
data if we try to sample directly from
using the rust package’s default settings for the generalized
ratio-of-uniforms algorithm. One solution is to reparameterize to
,
which hanova1 implements by default. Another possibility is
to increase the generalized ratio-of-uniforms tuning parameter
r from the default value of 1/2 used in rust.
These approaches are illustrated below.
coag1 <- hanova1(resp = coagulation[, 1], fac = coagulation[, 2], n = 10000)
coag2 <- hanova1(resp = coagulation[, 1], fac = coagulation[, 2], n = 10000,
param = "original", r = 1)
plot(coag1, params = "ru")
plot(coag1, ru_scale = TRUE)
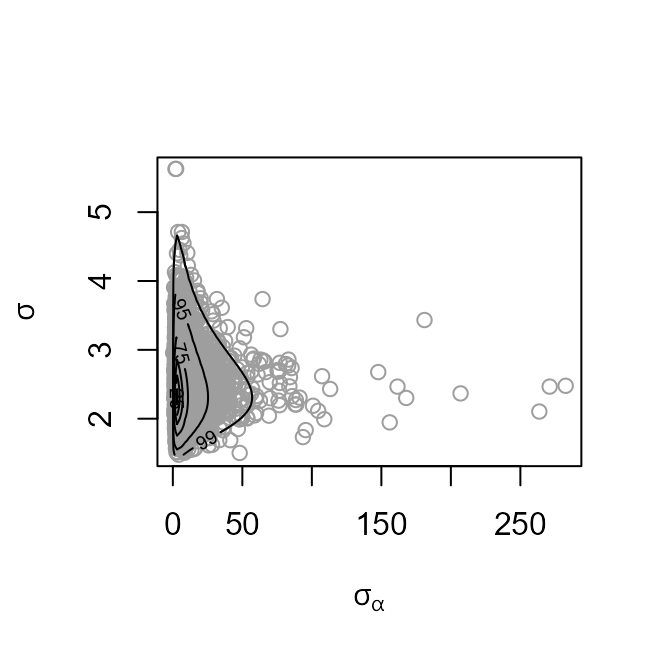
The heaviness of the upper tail of the marginal posterior density of
is evident in the plot on the left. Parameter transformation to
results in a density (in the plot on the right) from which it is easier
to simulate.
We produce some summaries of the posterior sample stored in
coag1. The summaries calculated from coag2 are
very similar.
probs <- c(2.5, 25, 50, 75, 97.5) / 100
all1 <- cbind(coag1$theta_sim_vals, coag1$sim_vals)
round(t(apply(all1, 2, quantile, probs = probs)), 1)
#> 2.5% 25% 50% 75% 97.5%
#> theta[1] 58.8 60.4 61.2 62.0 63.8
#> theta[2] 64.0 65.2 65.9 66.5 67.9
#> theta[3] 65.7 67.1 67.8 68.4 69.8
#> theta[4] 59.4 60.5 61.1 61.7 62.9
#> mu 54.7 62.2 64.0 65.7 73.2
#> sigma[alpha] 2.0 3.5 5.0 7.9 27.0
#> sigma 1.8 2.2 2.4 2.7 3.4
These posterior summaries are similar to those presented in Table
11.3 of Gelman et al. (2014) (where
is denoted
),
which were obtained using Gibbs sampling.
When the number of groups is small Gelman
(2006) advocates the use of a half-Cauchy prior for
.
The code below implements this using independent half-Cauchy priors for
and
,
that is,
We choose, somewhat arbitrarily,
:
in practice
should be set by considering the problem in hand. See Gelman (2006) for details. We set
to be large enough to result in an effectively flat prior for
.
coag3 <- hanova1(resp = coagulation[, 1], fac = coagulation[, 2],
param = "original", prior = "cauchy", hpars = c(10, 1e6))
Appendix
Consider the hierarchical 1-way ANOVA model
where
and
and all random variables are independent. We specify a prior density
for
.
Let
and
.
Marginal posterior density of
The joint posterior density of
satisfies
where
.
Let
.
Completing the square in
in the term in square brackets gives
where
and
.
Therefore,
The marginal posterior density of
is given by
Manipulating
gives
Therefore, the joint
posterior density of
satisfies
where
,
and
.
A special case
Suppose that a
)
prior distribution is specified for
,
so that
and that
and
are independent a priori. We derive the marginal posterior
density for
in this case and the conditional posterior density of
given
.
The joint posterior density for
satisfies
Completing the square in
in
gives
where
.
Therefore,
Retaining only the terms in
that involve
gives
Therefore,
.
The conditional posterior density of
given
The conditional posterior density of
given
satisfies
because these are the only
terms in
that involve
.
Therefore, conditional on
,
are independent a posteriori and
,
where
Noting that
gives
,
where
Factorisations for simulation
In the most general case considered here the factorisation
means that we can simulate
first from the three-dimensional
and then, conditional on the value of
,
simulate independently from each of the normal distributions of
.
In the special case detailed above the factorisation becomes
Therefore, the first stage
can be performed by simulating from the two-dimensional
and then, conditional on the value of
,
from the normal distribution of
.
References
Gelman, A. 2006.
“Prior Distributions for Variance Parameters in
Hierarchical Models.” Bayesian Analysis 1
(3): 515–33.
https://doi.org/10.1214/06-BA117A.
Gelman, A., J. B. Carlin, H. S. Stern, D. B. Dunson, A. Vehtari, and D.
B. Rubin. 2014.
Bayesian Data Analysis. Third. Florida, USA:
Chapman & Hall / CRC.
http://www.stat.columbia.edu/~gelman/book/.
Northrop, P. J. 2017.
rust:
Ratio-of-Uniforms Simulation with Transformation.
https://CRAN.R-project.org/package=rust.
Northrop, P. J., and R. E. Chandler. 2014.
“Quantifying Sources of
Uncertainty in Projections of Future Climate” 27: 8793–8808.
https://doi.org/10.1175/JCLI-D-14-00265.1.





