An overview of anscombiser
Paul Northrop
2026-01-02
Source:vignettes/intro-to-anscombiser.Rmd
intro-to-anscombiser.RmdThe anscombiser package is named after the famous
Anscombe’s quartet datasets (Anscombe
(1973)). Frank Anscombe created these datasets to emphasize the
importance of graphical techniques in statistical analyses. The datasets
each consist of 11 pairs of \((x, y)\)
points and they have almost exactly the same values of an apparently
remarkable number of sample summary statistics. However, scatter plots
of these data reveal that the behaviours exhibited in these datasets are
very different. That inspecting summary statistics can give a very
misleading impression of a dataset is an important point for students of
Statistics to appreciate.
Creating datasets with identical summary statistics
The datasauRus package (Locke and
D’Agostino McGowan (2018)) provides further examples of datasets
that have markedly different scatter plots but nevertheless share many
sample summary statistics. These datasets were produced by using a
simulated annealing algorithm that seeks to morph incrementally an
initial dataset towards a target shape while maintaining the same sample
summary statistics (Matejka and Fitzmaurice
(2017)). In principle, any set of summary statistics can be used.
Indeed, Locke and D’Agostino McGowan
(2018) provides not only datasets that have the same values of
Anscombe’s statistics (essentially sample means, variances and
correlation) but also datasets that are constrained to share the same
sample median, interquartile range and Spearman’s rank correlation.
The anscombiser package takes a simpler and quicker
approach to the same problem, using Anscombe’s statistics. It uses
shifting, scaling and rotating to transform the observations in an input
dataset to achieve a target set of Anscombe’s statistics. These
statistics can be set directly or by calculating them from a target
dataset, perhaps one of Anscombe’s quartet. If the input dataset has
statistics that are similar to the target statistics then the output
dataset will look rather similar to the input dataset. Otherwise, the
output dataset will be a squashed and/or rotated version of the input
dataset, but the general shape of the input dataset will still be
visible. It will be like viewing the input dataset from a different
perspective.
Thus, we can easily create many datasets that have different general natures but share the same values of Anscombe’s statistics. In addition, this method works in more than two dimensions.
Anscombe’s quartet
The anscombise() function takes an input two-dimensional
dataset and outputs a dataset that shares Anscombe’s statistics with his
quartet of datasets. The which argument chooses which of
Anscombe’s datasets to use. The default is which = 1. Of
course, this affects the output dataset only minimally but it matters if
we want to plot the input dataset, as we do in an example below.
The anscombiser packages provides 8 input files:
input1 to input8 that can be used to create
Anscombe-like datasets, with the same sample size of 11 as the original
Anscombe quartet. The following example uses input data arranged on the
edge of a circle.
a2 <- anscombise(input2, which = 4)
plot(a2)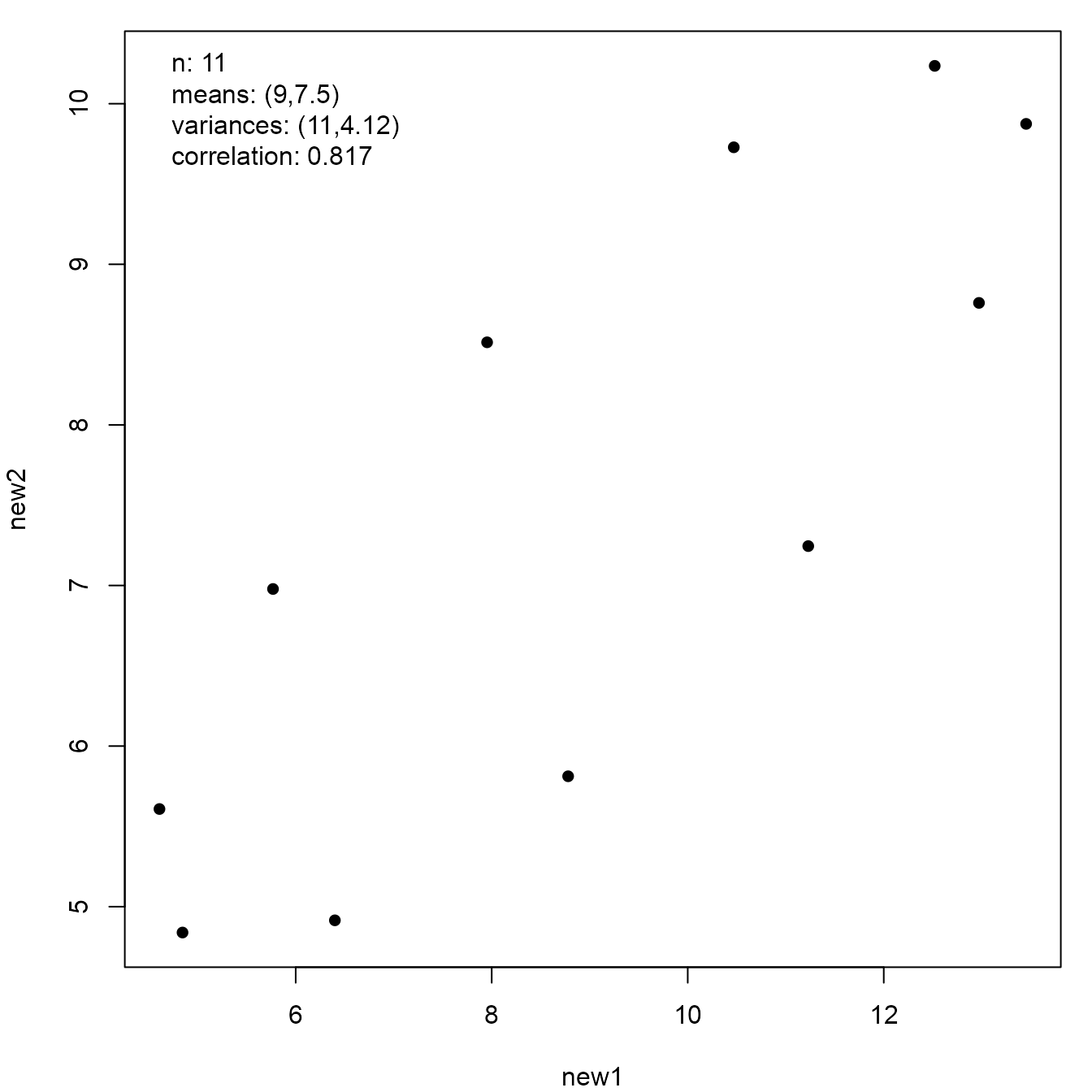
Now we transform the Old Faithful Geyser data so that it shares the sample summary statistics of the Anscombe quartet.
new_faithful <- anscombise(datasets::faithful, which = 4)
plot(new_faithful)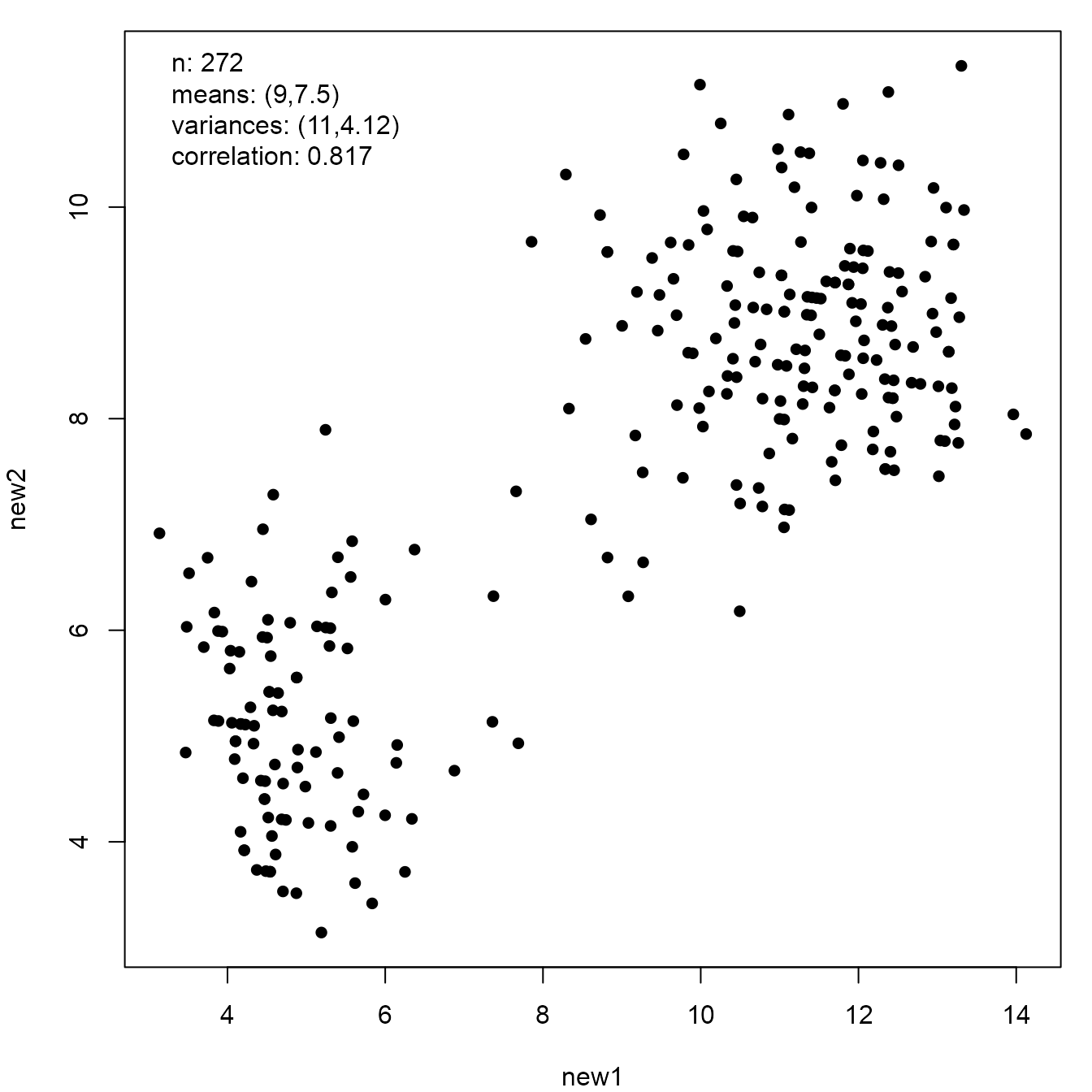
plot(new_faithful, input = TRUE)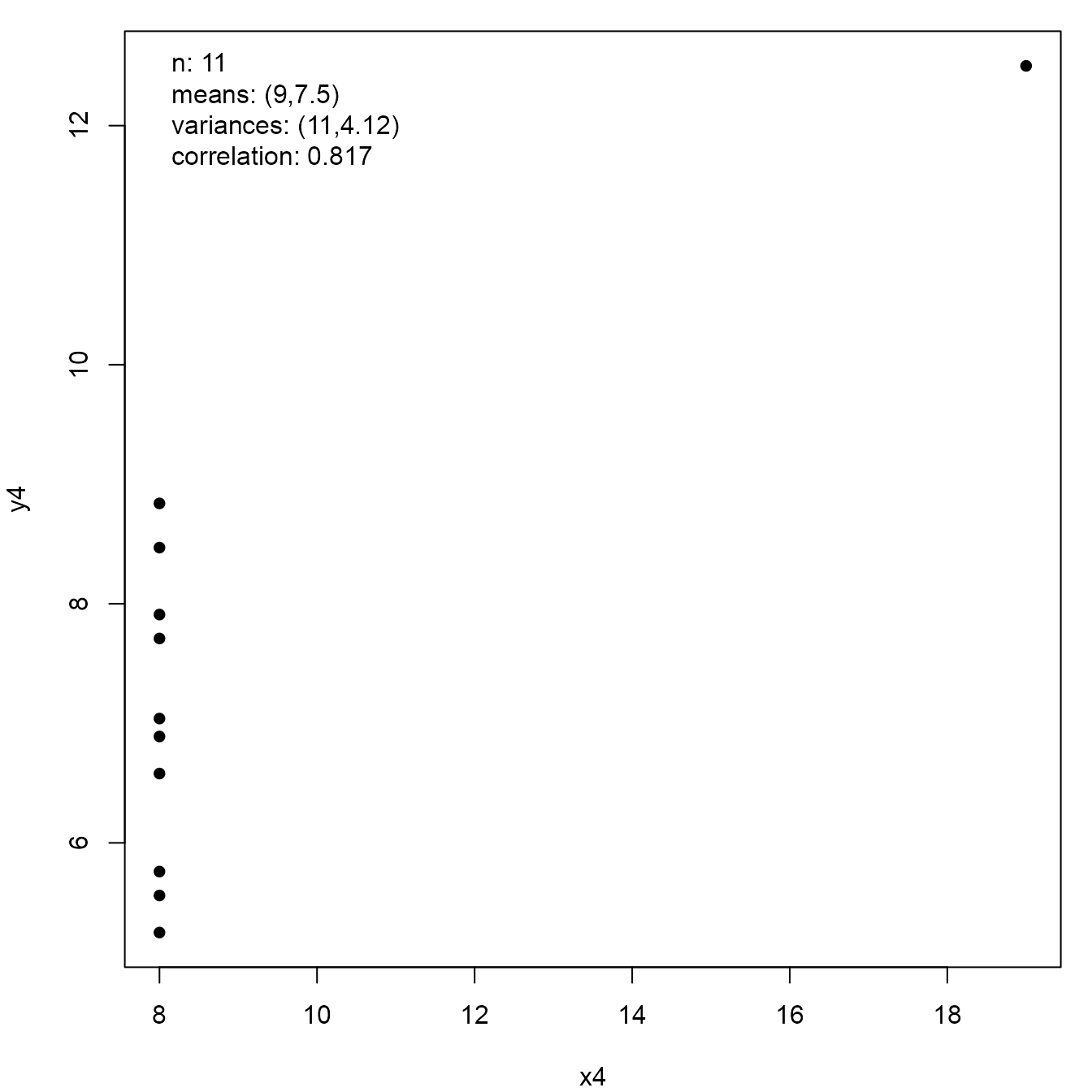
If we view a plot of the outline of the coast of Italy from a strange angle then the resulting dataset has the same sample summary statistics as those above.
italy <- mapdata("Italy")
new_italy <- anscombise(italy, which = 4)
plot(new_italy)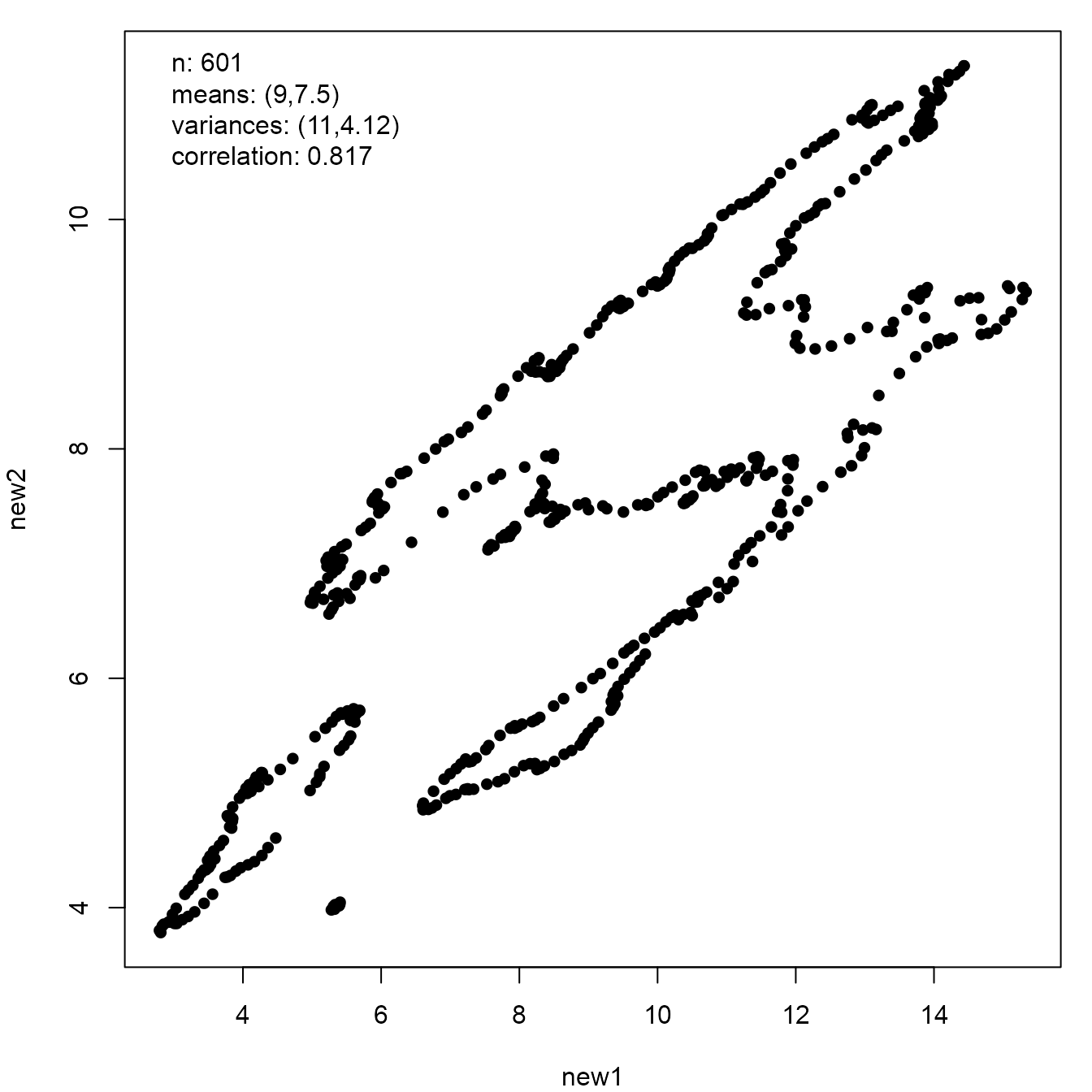
Other two-dimensional examples
The mimic() function of the anscombiser
package transforms an input dataset, as outlined above, to mimic another
dataset, in the sense of replicating its values of Anscombe’s
statistics. A particularly effective feature of the
datasauRus package is a dataset that draws a picture of a
dinosaur. Here, we show that a plot of the outline of the coast of the
UK needs little adjustment to replicate the sample summary statistics of
the dinosaur dataset.
library(datasauRus)
#> Warning: package 'datasauRus' was built under R version 4.5.2
library(maps)
#> Warning: package 'maps' was built under R version 4.5.2
dino <- datasaurus_dozen_wide[, c("dino_x", "dino_y")]
UK <- mapdata("UK")
new_UK <- mimic(UK, dino)
plot(new_UK, legend_args = list(x = "right"))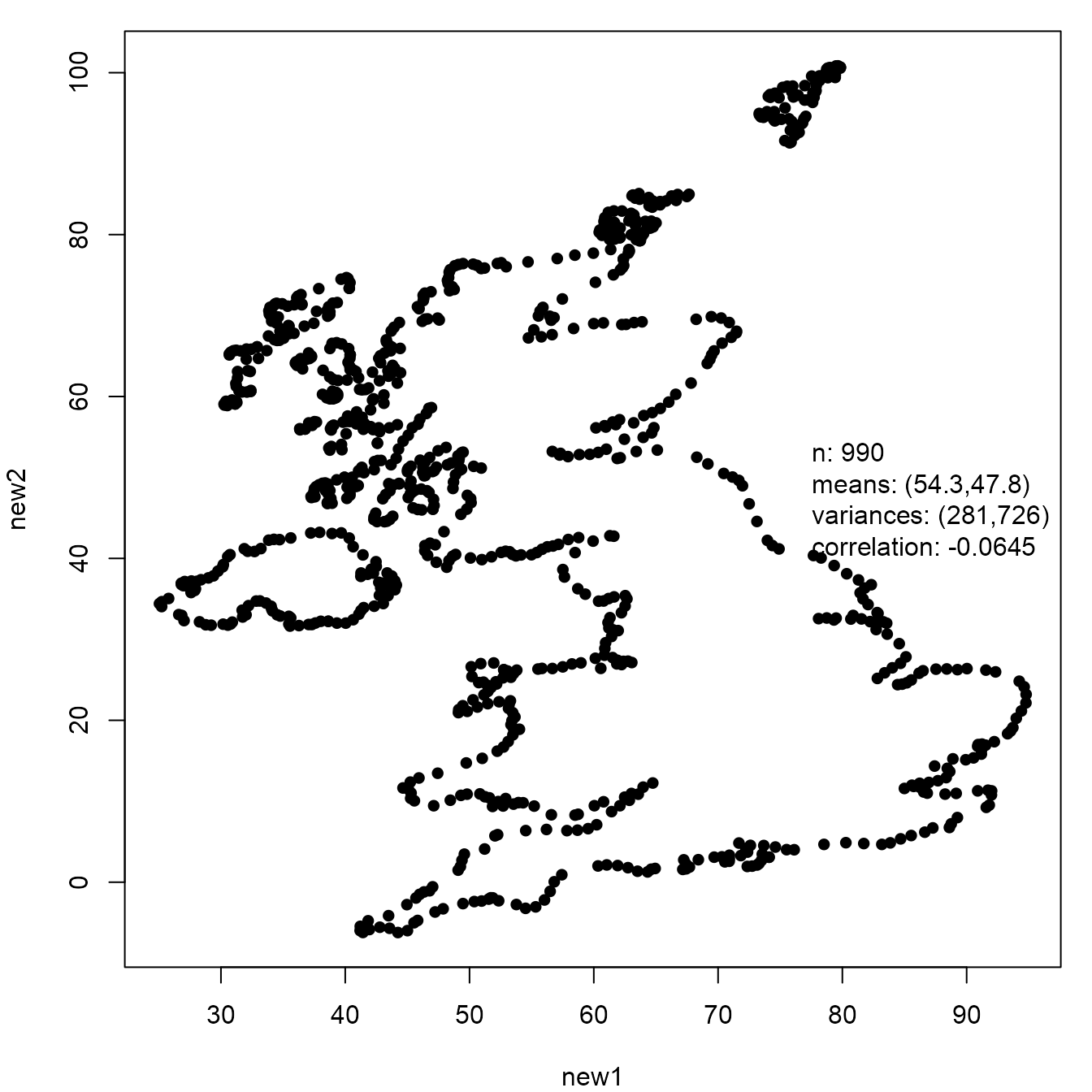
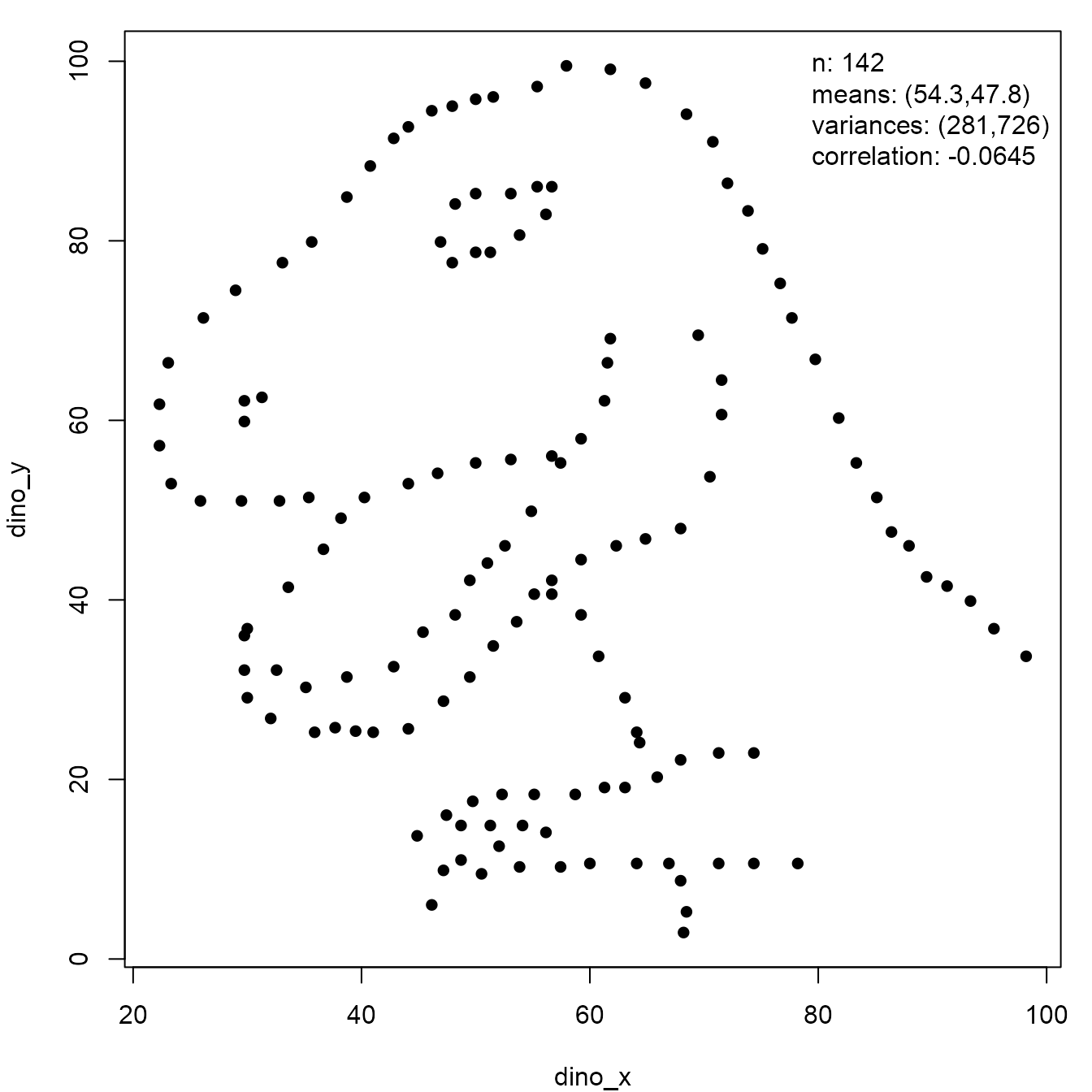
We finish this section with another example involving the dinosaur.
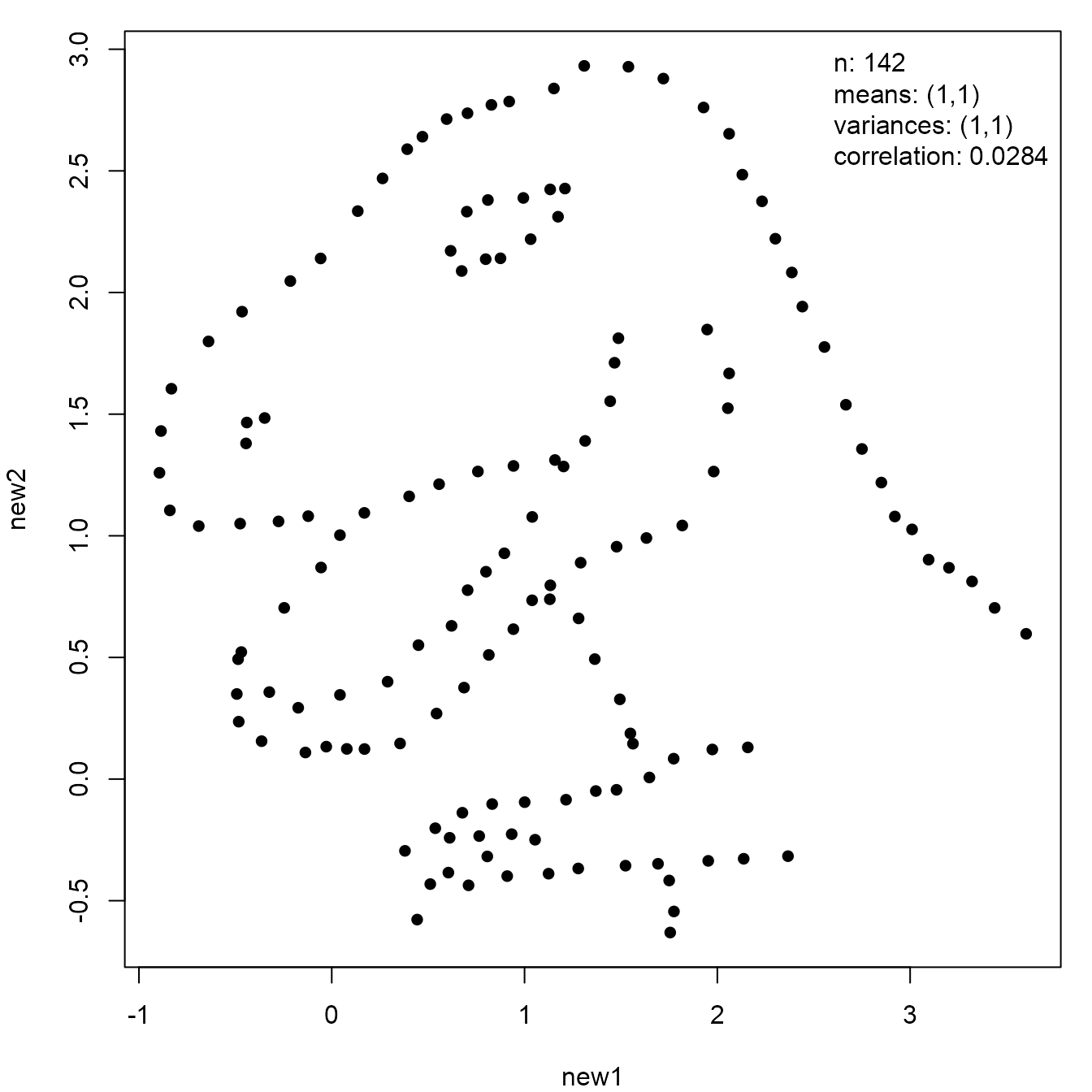
The final image was created by Accentaur from the Noun Project.
Three-dimensional examples
We conclude with a brief 3D example, using the randu and
trees datasets in the datasets package.
new_randu <- mimic(datasets::randu, datasets::trees)
# new_randu and trees share the same sample summary statistics
new_randu_stats <- get_stats(new_randu)
trees_stats <- get_stats(datasets::trees)
# For example
trees_stats$correlation
#> Girth Height Volume
#> Girth 1.0000000 0.5192801 0.9671194
#> Height 0.5192801 1.0000000 0.5982497
#> Volume 0.9671194 0.5982497 1.0000000
new_randu_stats$correlation
#> new1 new2 new3
#> new1 1.0000000 0.5192801 0.9671194
#> new2 0.5192801 1.0000000 0.5982497
#> new3 0.9671194 0.5982497 1.0000000
pairs(trees)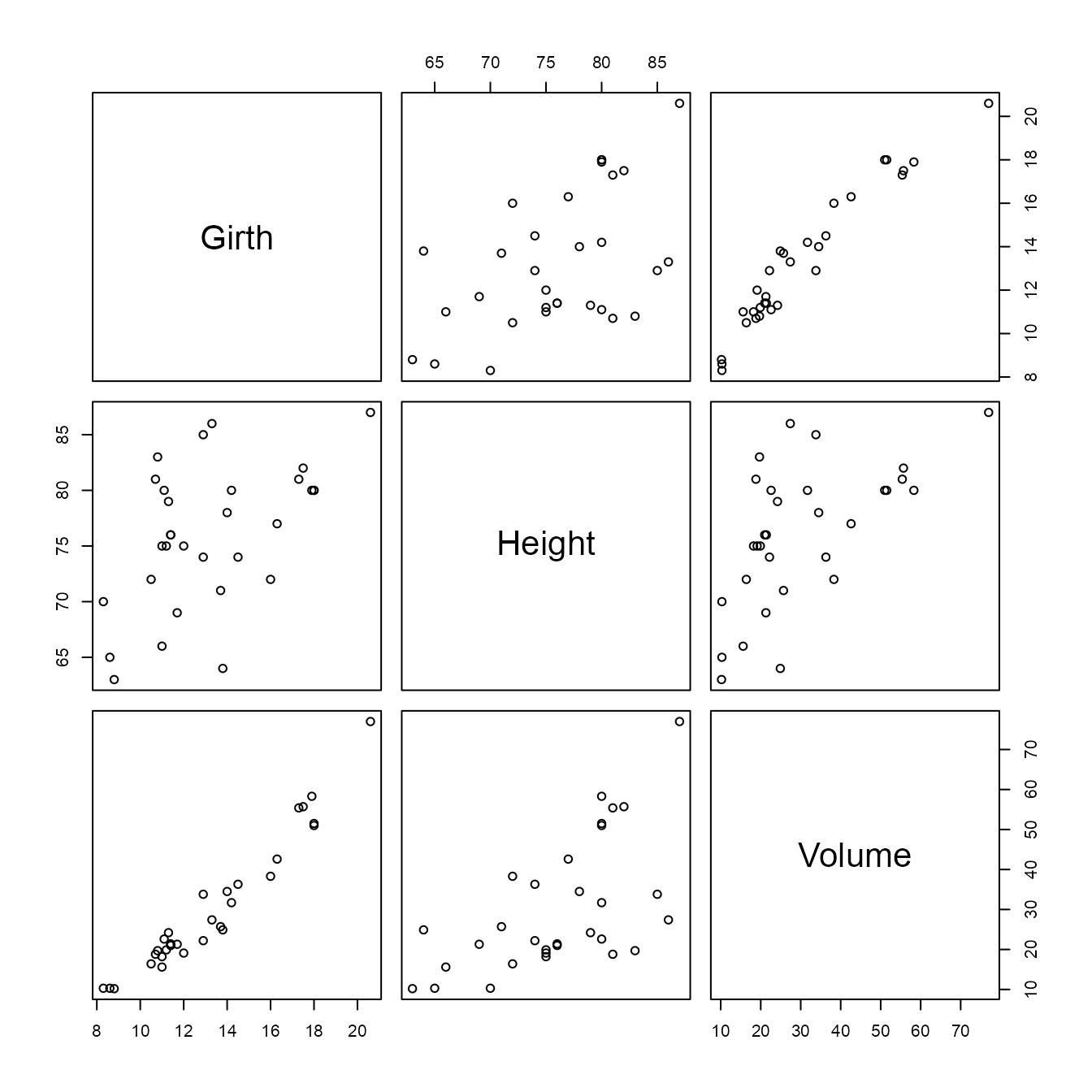
pairs(new_randu)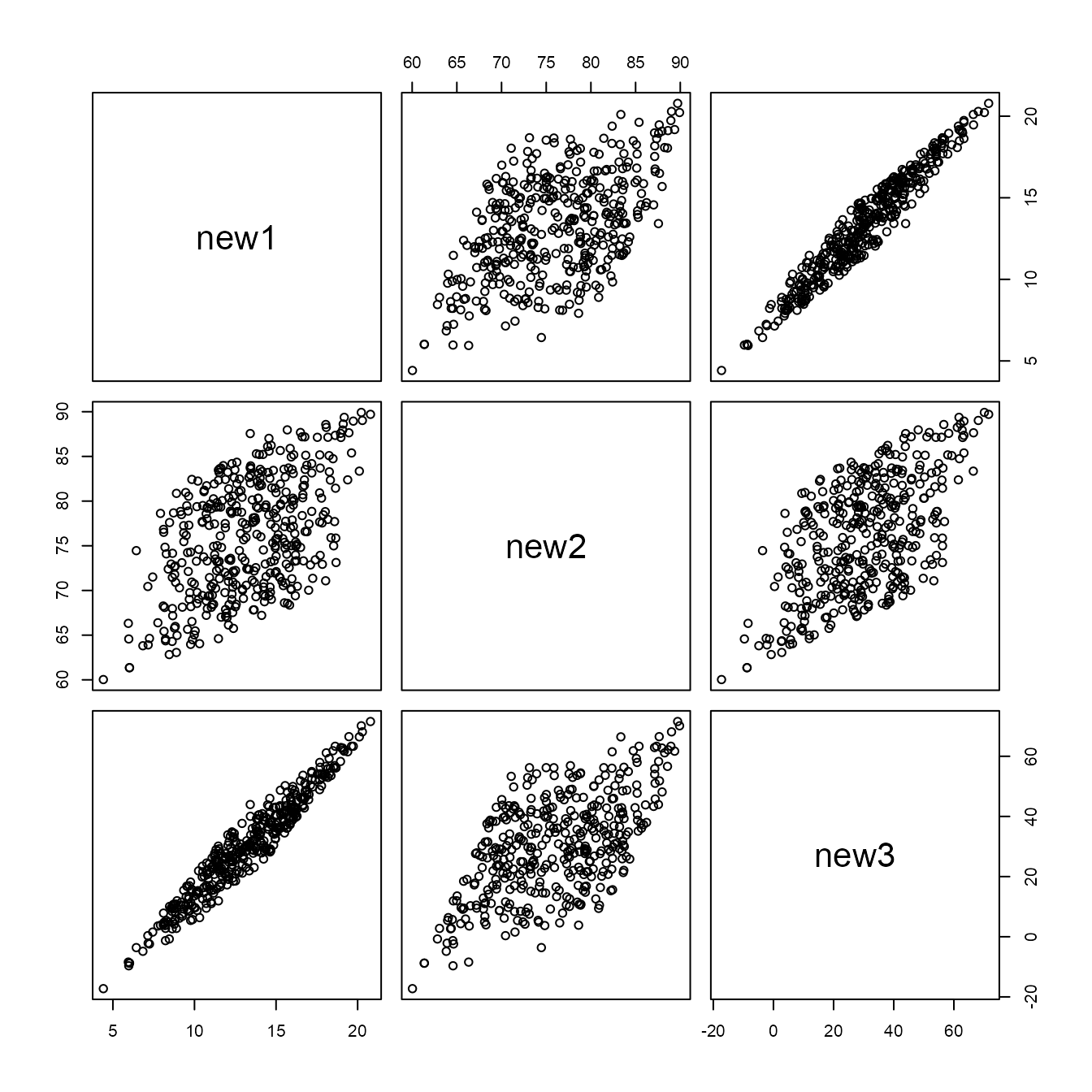
It is well-known that in three-dimensional displays of the
randu data non-random structure is evident, but this isn’t
evident in these pairwise displays.