Performs adjustments of a user-supplied independence loglikelihood for the presence of cluster dependence, following Chandler and Bate (2007). The user provides a function that returns a vector of observation-specific loglikelihood contributions and a vector that indicates cluster membership. The loglikelihood of a sub-model can be adjusted by fixing a set of parameters at particular values.
Usage
adjust_loglik(
loglik = NULL,
...,
cluster = NULL,
p = NULL,
init = NULL,
par_names = NULL,
fixed_pars = NULL,
fixed_at = 0,
name = NULL,
larger = NULL,
alg_deriv = NULL,
alg_hess = NULL,
mle = NULL,
H = NULL,
V = NULL
)Arguments
- loglik
A named function. Returns a vector of the loglikelihood contributions of individual observations. The first argument must be the vector of model parameter(s). If any of the model parameters are out-of-bounds then
loglikshould return either-Infor a vector with at least one element equal to-Inf. The number of parameters in the full model must be specified using (at least) one of the argumentsp,initorpar_names.- ...
Further arguments to be passed either to
loglik(and toalg_derivandalg_hessif these are supplied) or tooptim. The latter may includegr,method,lower,upperorcontrol. In the call tooptim,hessian = TRUEwill be used regardless of any value supplied. The functionloglikmust not have arguments with names that match any of these arguments tooptim.- cluster
A vector or factor indicating from which cluster the respective loglikelihood contributions from
loglikoriginate. Must have the same length as the vector returned byloglik. Ifclusteris not supplied then it is set insideadjust_loglikunder the assumption that each observation forms its own cluster.- p
A numeric scalar. The dimension of the full parameter vector, i.e. the number of parameters in the full model. Must be consistent with the lengths of
initandpar_names, if these are also supplied.- init
A numeric vector of initial values. Must have length equal to the number of parameters in the full model. If
initis supplied thenpis set tolength(init), provided that this is consistent with the the value given bypor implied bylength(par_names). Iffixed_parsis notNULLtheninit[-fixed_pars]is used in the search for the MLE. Ifinitis not supplied thenrep(0.1, p)is used.- par_names
A character vector. Names of the
pparameters in the full model. Must be consistent with the lengths ofinitandp, if these are also supplied.- fixed_pars
A vector specifying which parameters are to be restricted to be equal to the value(s) in
fixed_at. Can be either a numeric vector, specifying indices of the components of the full parameter vector, or a character vector of parameter names, which must be a subset of those supplied inpar_namesor stored in the objectlarger.- fixed_at
A numeric vector of length 1 or
length(fixed_pars). Iflength(fixed_at) = 1then the componentsfixed_parsof the parameter vector are all fixed atfixed_at. Iflength(fixed_at) = length(fixed_pars)then the componentfixed_pars[i]is fixed atfixed_at[i]for eachi.- name
A character scalar. A name for the model that gives rise to
loglik. If this is not supplied then the name inlargeris used, if this has been supplied, and the name of the functionloglikotherwise.- larger
Only relevant if
fixed_parsis notNULL. Iflargeris supplied butfixed_parsis not then an error will result. iflargeris supplied then information about the model inlarger, e.g. aboutpandpar_nameswill override any attempt to set these arguments in the call toadjust_loglik.An object of class
"chandwich"returned byadjust_loglik, corresponding to a model in which the smaller model implied byfixed_parsis nested. Iflargeris supplied then all the arguments toadjust_loglikapart fromfixed_parsandfixed_atare extracted fromlarger. Ifinitis not supplied in the current call toadjust_logliktheninitis set toattr(larger, "MLE"), with the elements infixed_parsset tofixed_at.- alg_deriv
A function with the vector of model parameter(s) as its first argument. Returns a
length(cluster)bypnumeric matrix. Column i contains the derivatives of each of the loglikelihood contributions inloglikwith respect to model parameter i.- alg_hess
A function with the vector of model parameter(s) as its first argument. Returns a
pbypnumeric matrix equal to the Hessian ofloglik, i.e. the matrix of second derivatives of the functionloglik.Supplying both
Vandalg_derivor bothHandalg_hesswill produce an error.- mle
A numeric vector. Can only be used if
fixed_pars = NULL. Provides the maximum likelihood estimate of the model parameters, that is, the value of the parameter vector at which the independence loglikelihoodloglikis maximized. Must have length equal to the number of parameters in the full model. Ifmleis supplied thenpis set tolength(mle), provided that this is consistent with the the value given bypor implied bylength(par_names). Ifmleis supplied then it overridesinit.- H, V
p by p numeric matrices. Only used if
mleis supplied. Provide estimates of the Hessian of the independence loglikelihood (H) and the variance of the vector of cluster-specific contributions to the score vector (first derivatives with respect to the parameters) of the independence loglikelihood, each evaluated at the MLEmle. See the Introducing chandwich vignette and/or Chandler and Bate (2007).Supplying both
Vandalg_derivor bothHandalg_hesswill produce an error.
Value
A function of class "chandwich" to evaluate an adjusted
loglikelihood, or the independence loglikelihood, at one or more sets
of model parameters, with arguments
- x
A numeric vector or matrix giving values of the
p_current(see below) parameters in the model to which the returned adjusted loglikelihood applies. Ifp_current = 1this may be a numeric vector or a matrix with 1 column. Ifp_current > 1this may be a numeric vector of lengthp(one set of model parameters) or a numeric matrix withpcolumns (nrow(x)sets of model parameters), one set in each row ofx.- type
A character scalar. The type of adjustment to use. One of
"vertical","cholesky","spectral"or"none".
The latter results in the evaluation of the (unadjusted) independence loglikelihood. The function has (additional) attributes
- p_full, p_current
The number of parameters in the full model and current models, respectively.
- free_pars
A numeric vector giving the indices of the free parameters in the current model, with names inferred from
par_namesif this was supplied.- MLE, res_MLE
Numeric vectors, with names inferred from
par_namesif this was supplied. Maximum likelihood estimates of free parameters under the current model (mle) and all parameters in the full model, including any parameters with fixed values (res_MLE).- SE, adjSE
The unadjusted and adjusted estimated standard errors, of the free parameters, respectively.
- VC, adjVC
The unadjusted and adjusted estimated variance-covariance matrix of the free parameters, respectively.
- HI, HA
The Hessians of the independence and adjusted loglikelihood, respectively.
- C_cholesky, C_spectral
The matrix C in equation (14) of Chandler and Bate (2007), calculated using Cholesky decomposition and spectral decomposition, respectively.
- full_par_names, par_names
The names of the parameters in the full and current models, respectively, if these were supplied in this call or a previous call.
- max_loglik
The common maximised value of the independence and adjusted loglikelihoods.
- loglik, cluster
The arguments
loglikandclustersupplied in this call, or a previous call.- loglik_args
A list containing the further arguments passed to
loglikvia ... in this call, or a previous call.- loglikVecMLE
a vector containing the contributions of individual observations to the independence log-likelihood evaluated at the MLE.
- name
The argument
name, or the name of the functionloglikifnameisn't supplied.- nobs
The number of observations.
- call
The call to
adjust_loglik.
If fixed_pars is not NULL then there are further attributes
- fixed_pars
The argument
fixed_pars, with names inferred frompar_namesif this was supplied.- fixed_at
The argument
fixed_at, with names inferred frompar_namesif this was supplied.
If alg_deriv and/or alg_hess were supplied then these are
returned as further attributes.
To view an individual attribute called att_name use
attr(x, "att_name") or attributes(x)$att_name.
Details
Three adjustments to the independence loglikelihood described in
Chandler and Bate (2007) are available. The vertical adjustment is
described in Section 6 and two horizontal adjustments are described
in Sections 3.2 to 3.4. See the descriptions of type and, for the
horizontal adjustments, the descriptions of C_cholesky and
C_spectral, in Value.
The adjustments involve first and second derivatives of the loglikelihood
with respect to the model parameters. These are estimated using
jacobian and optimHess
unless alg_deriv and/or alg_hess are supplied.
References
Chandler, R. E. and Bate, S. (2007). Inference for clustered data using the independence loglikelihood. Biometrika, 94(1), 167-183. doi:10.1093/biomet/asm015
See also
summary.chandwich for maximum likelihood estimates
and unadjusted and adjusted standard errors.
plot.chandwich for plots of one-dimensional adjusted
loglikelihoods.
confint.chandwich, anova.chandwich,
coef.chandwich, vcov.chandwich
and logLik.chandwich for other chandwich methods.
conf_intervals for confidence intervals for
individual parameters.
conf_region for a confidence region for
a pair of parameters.
compare_models to compare nested models using an
(adjusted) likelihood ratio test.
Examples
# ------------------------- Binomial model, rats data ----------------------
# Contributions to the independence loglikelihood
binom_loglik <- function(prob, data) {
if (prob < 0 || prob > 1) {
return(-Inf)
}
return(dbinom(data[, "y"], data[, "n"], prob, log = TRUE))
}
rat_res <- adjust_loglik(loglik = binom_loglik, data = rats, par_names = "p")
# Plot the loglikelihoods
plot(rat_res, type = 1:4, legend_pos = "bottom", lwd = 2, col = 1:4)
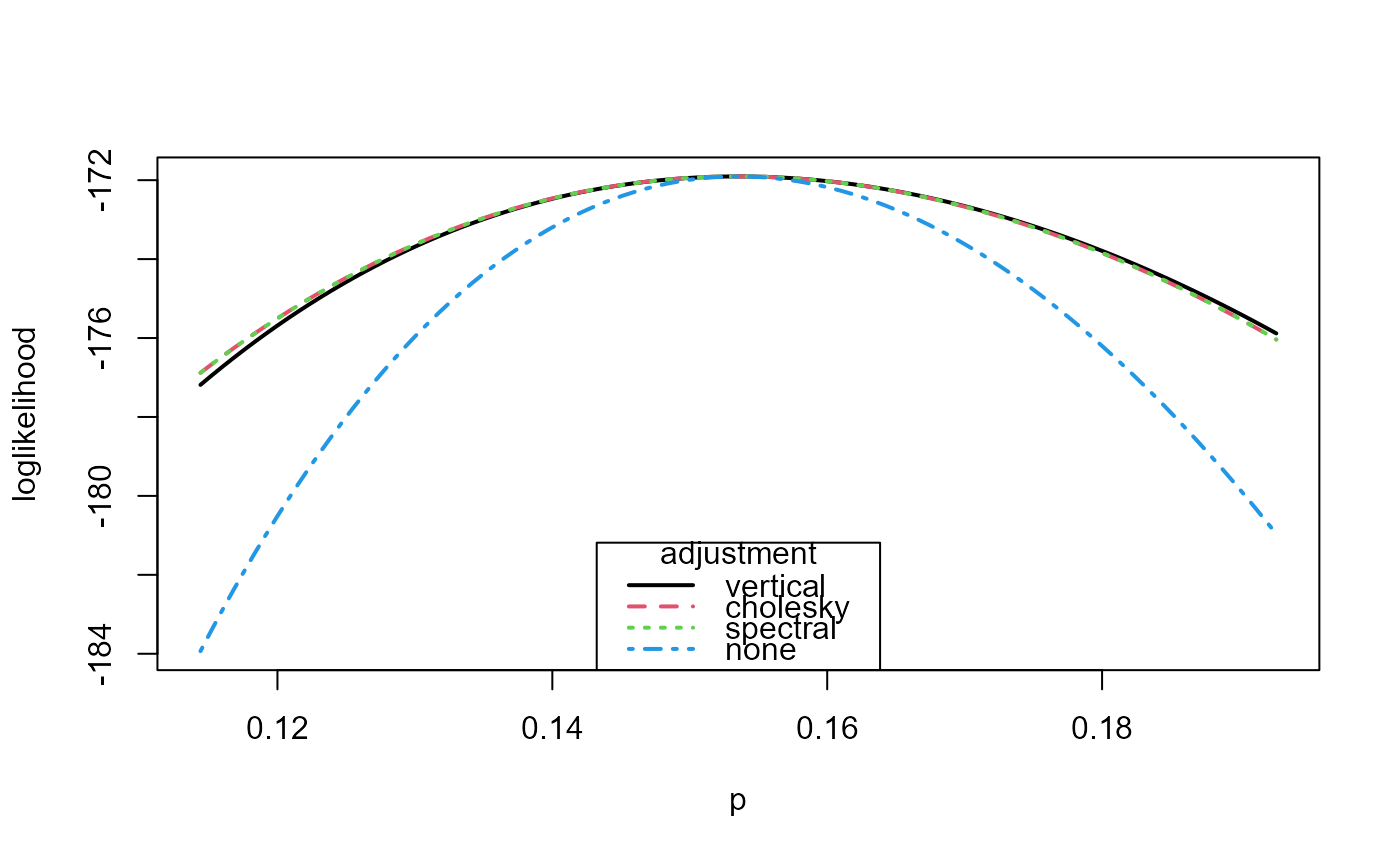 # MLE, SEs and adjusted SEs
summary(rat_res)
#> MLE SE adj. SE
#> p 0.1535 0.008645 0.01305
# -------------------------- GEV model, owtemps data -----------------------
# ------------ following Section 5.2 of Chandler and Bate (2007) -----------
# Contributions to the independence loglikelihood
gev_loglik <- function(pars, data) {
o_pars <- pars[c(1, 3, 5)] + pars[c(2, 4, 6)]
w_pars <- pars[c(1, 3, 5)] - pars[c(2, 4, 6)]
if (isTRUE(o_pars[2] <= 0 | w_pars[2] <= 0)) return(-Inf)
o_data <- data[, "Oxford"]
w_data <- data[, "Worthing"]
check <- 1 + o_pars[3] * (o_data - o_pars[1]) / o_pars[2]
if (isTRUE(any(check <= 0))) return(-Inf)
check <- 1 + w_pars[3] * (w_data - w_pars[1]) / w_pars[2]
if (isTRUE(any(check <= 0))) return(-Inf)
o_loglik <- log_gev(o_data, o_pars[1], o_pars[2], o_pars[3])
w_loglik <- log_gev(w_data, w_pars[1], w_pars[2], w_pars[3])
return(o_loglik + w_loglik)
}
# Initial estimates (method of moments for the Gumbel case)
sigma <- as.numeric(sqrt(6 * diag(var(owtemps))) / pi)
mu <- as.numeric(colMeans(owtemps) - 0.57722 * sigma)
init <- c(mean(mu), -diff(mu) / 2, mean(sigma), -diff(sigma) / 2, 0, 0)
# Loglikelihood adjustment for the full model
par_names <- c("mu[0]", "mu[1]", "sigma[0]", "sigma[1]", "xi[0]", "xi[1]")
large <- adjust_loglik(gev_loglik, data = owtemps, init = init,
par_names = par_names)
# Rows 1, 3 and 4 of Table 2 of Chandler and Bate (2007)
t(summary(large))
#> mu[0] mu[1] sigma[0] sigma[1] xi[0] xi[1]
#> MLE 81.1700 2.6680 3.7290 0.5312 -0.19890 -0.08835
#> SE 0.3282 0.3282 0.2293 0.2293 0.04937 0.04937
#> adj. SE 0.4036 0.2128 0.2426 0.1911 0.03943 0.03625
# Loglikelihood adjustment of some smaller models: xi[1] = 0 etc
# Starting from a larger model
medium <- adjust_loglik(larger = large, fixed_pars = "xi[1]")
small <- adjust_loglik(larger = large, fixed_pars = c("sigma[1]", "xi[1]"))
small <- adjust_loglik(larger = medium, fixed_pars = c("sigma[1]", "xi[1]"))
# Starting from scratch
medium <- adjust_loglik(gev_loglik, data = owtemps, init = init,
par_names = par_names, fixed_pars = "xi[1]")
small <- adjust_loglik(gev_loglik, data = owtemps, init = init,
par_names = par_names, fixed_pars = c("sigma[1]", "xi[1]"))
# --------- Misspecified Poisson model for negative binomial data ----------
# ... following Section 5.1 of the "Object-Oriented Computation of Sandwich
# Estimators" vignette of the sandwich package
# https://cran.r-project.org/web/packages/sandwich/vignettes/sandwich-OOP.pdf
# Simulate data
set.seed(123)
x <- rnorm(250)
y <- rnbinom(250, mu = exp(1 + x), size = 1)
# Fit misspecified Poisson model
fm_pois <- glm(y ~ x + I(x^2), family = poisson)
summary(fm_pois)$coefficients
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 1.06326821 0.04135723 25.709367 9.184267e-146
#> x 0.99607219 0.05353446 18.606186 2.862861e-77
#> I(x^2) -0.04912373 0.02314608 -2.122335 3.380961e-02
# Contributions to the independence loglikelihood
pois_glm_loglik <- function(pars, y, x) {
log_mu <- pars[1] + pars[2] * x + pars[3] * x ^ 2
return(dpois(y, lambda = exp(log_mu), log = TRUE))
}
pars <- c("alpha", "beta", "gamma")
pois_quad <- adjust_loglik(pois_glm_loglik, y = y, x = x, par_names = pars)
summary(pois_quad)
#> MLE SE adj. SE
#> alpha 1.06300 0.04136 0.08378
#> beta 0.99610 0.05354 0.10520
#> gamma -0.04913 0.02315 0.03628
# Providing algebraic derivatives and Hessian
pois_alg_deriv <- function(pars, y, x) {
mu <- exp(pars[1] + pars[2] * x + pars[3] * x ^ 2)
return(cbind(y - mu, x * (y - mu), x ^2 * (y - mu)))
}
pois_alg_hess <- function(pars, y, x) {
mu <- exp(pars[1] + pars[2] * x + pars[3] * x ^ 2)
alg_hess <- matrix(0, 3, 3)
alg_hess[1, ] <- -c(sum(mu), sum(x * mu), sum(x ^ 2 * mu))
alg_hess[2, ] <- -c(sum(x * mu), sum(x ^ 2 * mu), sum(x ^ 3 * mu))
alg_hess[3, ] <- -c(sum(x ^ 2 * mu), sum(x ^ 3 * mu), sum(x ^ 4 * mu))
return(alg_hess)
}
pois_quad <- adjust_loglik(pois_glm_loglik, y = y, x = x, p = 3,
alg_deriv = pois_alg_deriv, alg_hess = pois_alg_hess)
summary(pois_quad)
#> MLE SE adj. SE
#> 1 1.06300 0.04136 0.08378
#> 2 0.99610 0.05354 0.10520
#> 3 -0.04913 0.02315 0.03628
got_sandwich <- requireNamespace("sandwich", quietly = TRUE)
if (got_sandwich) {
# Providing MLE, H and V
# H and V calculated using bread() and meat() from sandwich package
n_obs <- stats::nobs(fm_pois)
pois_quad <- adjust_loglik(pois_glm_loglik, y = y, x = x, p = 3,
mle = fm_pois$coefficients,
H = -solve(sandwich::bread(fm_pois) / n_obs),
V = sandwich::meat(fm_pois) * n_obs)
}
# MLE, SEs and adjusted SEs
summary(rat_res)
#> MLE SE adj. SE
#> p 0.1535 0.008645 0.01305
# -------------------------- GEV model, owtemps data -----------------------
# ------------ following Section 5.2 of Chandler and Bate (2007) -----------
# Contributions to the independence loglikelihood
gev_loglik <- function(pars, data) {
o_pars <- pars[c(1, 3, 5)] + pars[c(2, 4, 6)]
w_pars <- pars[c(1, 3, 5)] - pars[c(2, 4, 6)]
if (isTRUE(o_pars[2] <= 0 | w_pars[2] <= 0)) return(-Inf)
o_data <- data[, "Oxford"]
w_data <- data[, "Worthing"]
check <- 1 + o_pars[3] * (o_data - o_pars[1]) / o_pars[2]
if (isTRUE(any(check <= 0))) return(-Inf)
check <- 1 + w_pars[3] * (w_data - w_pars[1]) / w_pars[2]
if (isTRUE(any(check <= 0))) return(-Inf)
o_loglik <- log_gev(o_data, o_pars[1], o_pars[2], o_pars[3])
w_loglik <- log_gev(w_data, w_pars[1], w_pars[2], w_pars[3])
return(o_loglik + w_loglik)
}
# Initial estimates (method of moments for the Gumbel case)
sigma <- as.numeric(sqrt(6 * diag(var(owtemps))) / pi)
mu <- as.numeric(colMeans(owtemps) - 0.57722 * sigma)
init <- c(mean(mu), -diff(mu) / 2, mean(sigma), -diff(sigma) / 2, 0, 0)
# Loglikelihood adjustment for the full model
par_names <- c("mu[0]", "mu[1]", "sigma[0]", "sigma[1]", "xi[0]", "xi[1]")
large <- adjust_loglik(gev_loglik, data = owtemps, init = init,
par_names = par_names)
# Rows 1, 3 and 4 of Table 2 of Chandler and Bate (2007)
t(summary(large))
#> mu[0] mu[1] sigma[0] sigma[1] xi[0] xi[1]
#> MLE 81.1700 2.6680 3.7290 0.5312 -0.19890 -0.08835
#> SE 0.3282 0.3282 0.2293 0.2293 0.04937 0.04937
#> adj. SE 0.4036 0.2128 0.2426 0.1911 0.03943 0.03625
# Loglikelihood adjustment of some smaller models: xi[1] = 0 etc
# Starting from a larger model
medium <- adjust_loglik(larger = large, fixed_pars = "xi[1]")
small <- adjust_loglik(larger = large, fixed_pars = c("sigma[1]", "xi[1]"))
small <- adjust_loglik(larger = medium, fixed_pars = c("sigma[1]", "xi[1]"))
# Starting from scratch
medium <- adjust_loglik(gev_loglik, data = owtemps, init = init,
par_names = par_names, fixed_pars = "xi[1]")
small <- adjust_loglik(gev_loglik, data = owtemps, init = init,
par_names = par_names, fixed_pars = c("sigma[1]", "xi[1]"))
# --------- Misspecified Poisson model for negative binomial data ----------
# ... following Section 5.1 of the "Object-Oriented Computation of Sandwich
# Estimators" vignette of the sandwich package
# https://cran.r-project.org/web/packages/sandwich/vignettes/sandwich-OOP.pdf
# Simulate data
set.seed(123)
x <- rnorm(250)
y <- rnbinom(250, mu = exp(1 + x), size = 1)
# Fit misspecified Poisson model
fm_pois <- glm(y ~ x + I(x^2), family = poisson)
summary(fm_pois)$coefficients
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 1.06326821 0.04135723 25.709367 9.184267e-146
#> x 0.99607219 0.05353446 18.606186 2.862861e-77
#> I(x^2) -0.04912373 0.02314608 -2.122335 3.380961e-02
# Contributions to the independence loglikelihood
pois_glm_loglik <- function(pars, y, x) {
log_mu <- pars[1] + pars[2] * x + pars[3] * x ^ 2
return(dpois(y, lambda = exp(log_mu), log = TRUE))
}
pars <- c("alpha", "beta", "gamma")
pois_quad <- adjust_loglik(pois_glm_loglik, y = y, x = x, par_names = pars)
summary(pois_quad)
#> MLE SE adj. SE
#> alpha 1.06300 0.04136 0.08378
#> beta 0.99610 0.05354 0.10520
#> gamma -0.04913 0.02315 0.03628
# Providing algebraic derivatives and Hessian
pois_alg_deriv <- function(pars, y, x) {
mu <- exp(pars[1] + pars[2] * x + pars[3] * x ^ 2)
return(cbind(y - mu, x * (y - mu), x ^2 * (y - mu)))
}
pois_alg_hess <- function(pars, y, x) {
mu <- exp(pars[1] + pars[2] * x + pars[3] * x ^ 2)
alg_hess <- matrix(0, 3, 3)
alg_hess[1, ] <- -c(sum(mu), sum(x * mu), sum(x ^ 2 * mu))
alg_hess[2, ] <- -c(sum(x * mu), sum(x ^ 2 * mu), sum(x ^ 3 * mu))
alg_hess[3, ] <- -c(sum(x ^ 2 * mu), sum(x ^ 3 * mu), sum(x ^ 4 * mu))
return(alg_hess)
}
pois_quad <- adjust_loglik(pois_glm_loglik, y = y, x = x, p = 3,
alg_deriv = pois_alg_deriv, alg_hess = pois_alg_hess)
summary(pois_quad)
#> MLE SE adj. SE
#> 1 1.06300 0.04136 0.08378
#> 2 0.99610 0.05354 0.10520
#> 3 -0.04913 0.02315 0.03628
got_sandwich <- requireNamespace("sandwich", quietly = TRUE)
if (got_sandwich) {
# Providing MLE, H and V
# H and V calculated using bread() and meat() from sandwich package
n_obs <- stats::nobs(fm_pois)
pois_quad <- adjust_loglik(pois_glm_loglik, y = y, x = x, p = 3,
mle = fm_pois$coefficients,
H = -solve(sandwich::bread(fm_pois) / n_obs),
V = sandwich::meat(fm_pois) * n_obs)
}