Calculates confidence intervals for individual parameters.
Usage
conf_intervals(
object,
which_pars = NULL,
init = NULL,
conf = 95,
mult = 1.5,
num = 10,
type = c("vertical", "cholesky", "spectral", "none"),
profile = TRUE,
...
)Arguments
- object
An object of class
"chandwich"returned byadjust_loglik.- which_pars
A vector specifying the (unfixed) parameters for which confidence intervals are required. Can be either a numeric vector, specifying indices of the components of the full parameter vector, or a character vector of parameter names, which must be a subset of those supplied in
par_namesin the call toadjust_loglikthat producedobject.which_parsmust not have any parameters in common withattr(object, "fixed_pars").which_parsmust not contain all of the unfixed parameters, i.e. there is no point in profiling over all the unfixed parameters.If missing, all parameters are included.
- init
A numeric vector of initial estimates of the values of the parameters that are not fixed and are not in
which_pars. Should have lengthattr(object, "p_current") - length(which_pars). IfinitisNULLor is of the wrong length then the relevant components from the MLE stored inobjectare used.- conf
A numeric scalar in (0, 100). Confidence level for the intervals.
- mult
A numeric vector of length 1 or the same length as
which_pars. The search for the profile loglikelihood-based confidence limits is conducted over the corresponding symmetric confidence intervals (based on approximate normal theory), extended by a factor of the corresponding component ofmult.- num
A numeric scalar. The number of values at which to evaluate the profile loglikelihood either side of the MLE. Increasing
numincreases the accuracy of the confidence limits, but the code will take longer to run.- type
A character scalar. The argument
typeto the function returned byadjust_loglik, that is, the type of adjustment made to the independence loglikelihood function.- profile
A logical scalar. If
FALSEthen only intervals based on approximate large sample normal theory, which are symmetric about the MLE, are returned (insym_CI) andprof_CIin the returned object will containNAs.- ...
Further arguments to be passed to
optim. These may includegr,method,lower,upperorcontrol.
Value
An object of class "confint", a list with components
- conf
The argument
conf.- cutoff
A numeric scalar. For values inside the confidence interval the profile loglikelihood lies above
cutoff.- parameter_vals, prof_loglik_vals
2 * num + 1bylength(which_pars)numeric matrices. Column i ofparameter_valscontains the profiled values of parameterwhich_par[i]. Column i ofprof_loglik_valscontains the corresponding values of the profile loglikelihood.- sym_CI, prof_CI
length(which_pars)by 2 numeric matrices. Row i ofsym_CI(prof_CI) contains the symmetric (profile loglikelihood-based) confidence intervals for parameterwhich_pars[i].
If a value in
prof_CI is NA then this means that the search for the
confidence limit did no extend far enough. A remedy is to increase
the value of mult, or the relevant component of mult,
and perhaps also increase num.
- max_loglik
The value of the adjusted loglikelihood at its maximum, stored in
object$max_loglik.- type
The argument
typesupplied in the call toconf_intervals, i.e. the type of loglikelihood adjustment.- which_pars
The argument
which_pars.- name
A character scalar. The name of the model, stored in
attr(object, "name").- p_current
The number of free parameters in the current model.
- fixed_pars, fixed_at
attr(object, "fixed_pars")andattr(object, "fixed_at"), the argumentsfixed_parsandfixed_attoadjust_loglik, if these were supplied.
Details
Calculates (profile, if necessary) likelihood-based confidence
intervals for individual parameters, and also provides symmetric intervals
based on a normal approximation to the sampling distribution of the
estimator. See also the S3 confint method
confint.chandwich.
See also
confint.chandwich S3 confint method for objects
of class "chandwich" returned from adjust_loglik.
adjust_loglik to adjust a user-supplied
loglikelihood function.
summary.chandwich for maximum likelihood estimates
and unadjusted and adjusted standard errors.
plot.chandwich for plots of one-dimensional adjusted
loglikelihoods.
conf_region for a confidence region for
a pair of parameters.
compare_models to compare nested models using an
(adjusted) likelihood ratio test.
Examples
# ------------------------- Binomial model, rats data ----------------------
# Contributions to the independence loglikelihood
binom_loglik <- function(prob, data) {
if (prob < 0 || prob > 1) {
return(-Inf)
}
return(dbinom(data[, "y"], data[, "n"], prob, log = TRUE))
}
rat_res <- adjust_loglik(loglik = binom_loglik, data = rats, par_names = "p")
# 95% likelihood-based confidence intervals, vertically adjusted
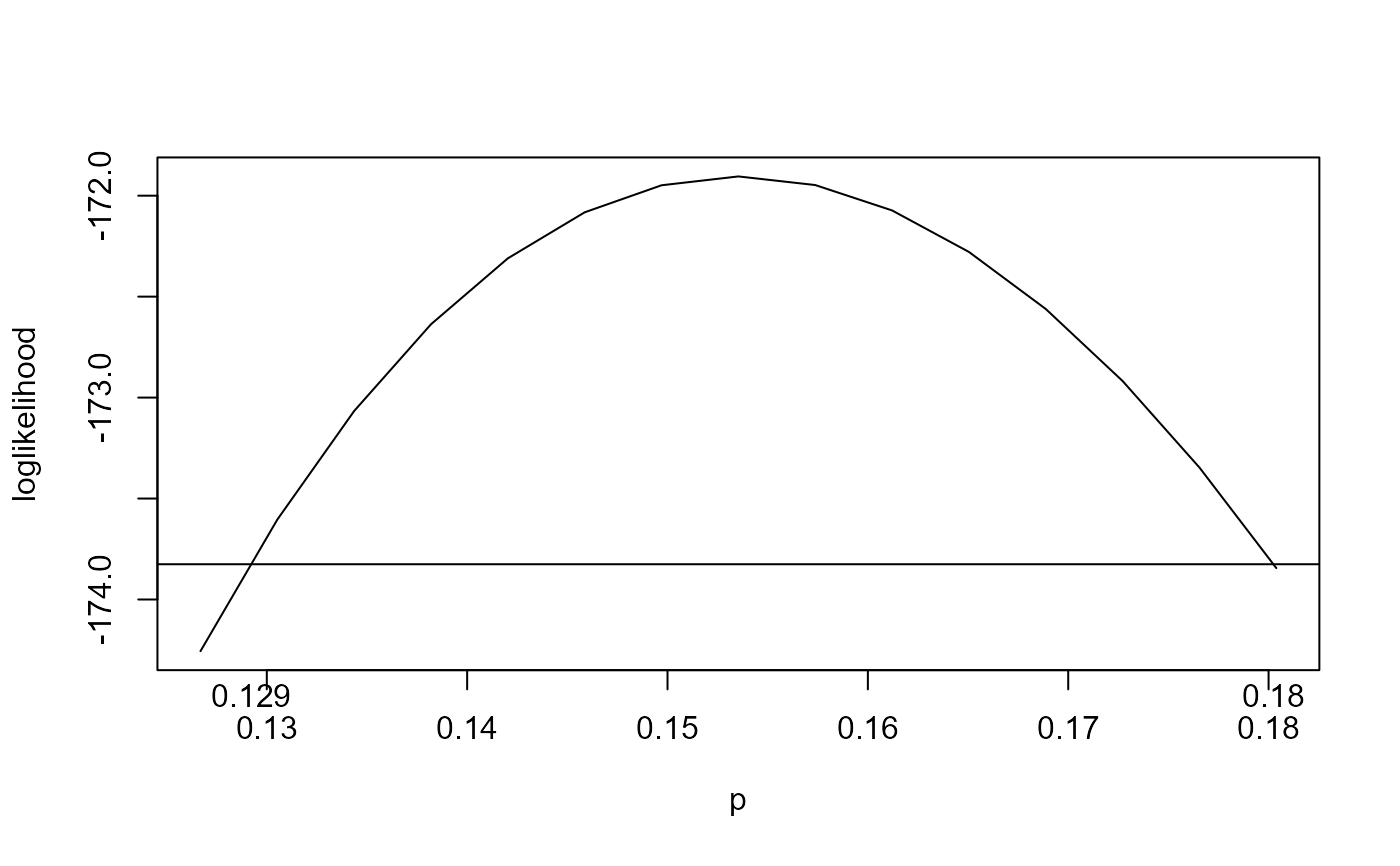
ci <- conf_intervals(rat_res)
#> Waiting for profiling to be done...
plot(ci)
 # Unadjusted
conf_intervals(rat_res, type = "none")
#> Waiting for profiling to be done...
#> Model: binom_loglik
#>
#> 95% confidence interval, independence loglikelihood
#>
#> Symmetric:
#> lower upper
#> p 0.1366 0.1705
#>
#> Likelihood-based:
#> lower upper
#> p 0.1372 0.1710
# -------------------------- GEV model, owtemps data -----------------------
# ------------ following Section 5.2 of Chandler and Bate (2007) -----------
gev_loglik <- function(pars, data) {
o_pars <- pars[c(1, 3, 5)] + pars[c(2, 4, 6)]
w_pars <- pars[c(1, 3, 5)] - pars[c(2, 4, 6)]
if (isTRUE(o_pars[2] <= 0 | w_pars[2] <= 0)) return(-Inf)
o_data <- data[, "Oxford"]
w_data <- data[, "Worthing"]
check <- 1 + o_pars[3] * (o_data - o_pars[1]) / o_pars[2]
if (isTRUE(any(check <= 0))) return(-Inf)
check <- 1 + w_pars[3] * (w_data - w_pars[1]) / w_pars[2]
if (isTRUE(any(check <= 0))) return(-Inf)
o_loglik <- log_gev(o_data, o_pars[1], o_pars[2], o_pars[3])
w_loglik <- log_gev(w_data, w_pars[1], w_pars[2], w_pars[3])
return(o_loglik + w_loglik)
}
# Initial estimates (method of moments for the Gumbel case)
sigma <- as.numeric(sqrt(6 * diag(var(owtemps))) / pi)
mu <- as.numeric(colMeans(owtemps) - 0.57722 * sigma)
init <- c(mean(mu), -diff(mu) / 2, mean(sigma), -diff(sigma) / 2, 0, 0)
# Log-likelihood adjustment of the full model
par_names <- c("mu[0]", "mu[1]", "sigma[0]", "sigma[1]", "xi[0]", "xi[1]")
large <- adjust_loglik(gev_loglik, data = owtemps, init = init,
par_names = par_names)
# 95% likelihood-based confidence intervals, vertically adjusted
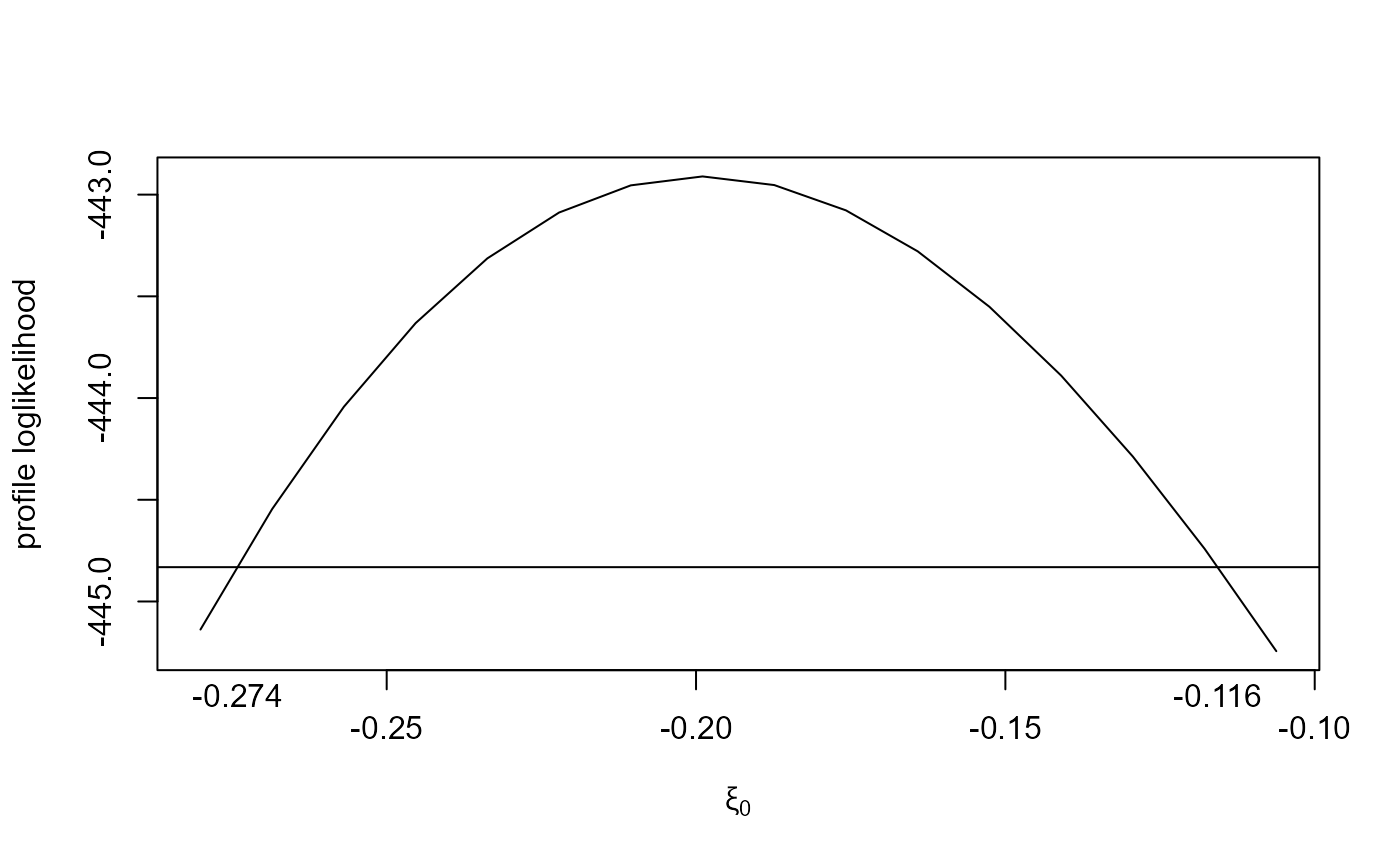
large_v <- conf_intervals(large, which_pars = c("xi[0]", "xi[1]"))
#> Waiting for profiling to be done...
large_v
#> Model: gev_loglik
#>
#> 95% confidence intervals, adjusted loglikelihod with type = ''vertical''
#>
#> Symmetric:
#> lower upper
#> xi[0] -0.27623 -0.12166
#> xi[1] -0.15939 -0.01731
#>
#> Profile likelihood-based:
#> lower upper
#> xi[0] -0.27410 -0.11572
#> xi[1] -0.16519 -0.02002
plot(large_v)
# Unadjusted
conf_intervals(rat_res, type = "none")
#> Waiting for profiling to be done...
#> Model: binom_loglik
#>
#> 95% confidence interval, independence loglikelihood
#>
#> Symmetric:
#> lower upper
#> p 0.1366 0.1705
#>
#> Likelihood-based:
#> lower upper
#> p 0.1372 0.1710
# -------------------------- GEV model, owtemps data -----------------------
# ------------ following Section 5.2 of Chandler and Bate (2007) -----------
gev_loglik <- function(pars, data) {
o_pars <- pars[c(1, 3, 5)] + pars[c(2, 4, 6)]
w_pars <- pars[c(1, 3, 5)] - pars[c(2, 4, 6)]
if (isTRUE(o_pars[2] <= 0 | w_pars[2] <= 0)) return(-Inf)
o_data <- data[, "Oxford"]
w_data <- data[, "Worthing"]
check <- 1 + o_pars[3] * (o_data - o_pars[1]) / o_pars[2]
if (isTRUE(any(check <= 0))) return(-Inf)
check <- 1 + w_pars[3] * (w_data - w_pars[1]) / w_pars[2]
if (isTRUE(any(check <= 0))) return(-Inf)
o_loglik <- log_gev(o_data, o_pars[1], o_pars[2], o_pars[3])
w_loglik <- log_gev(w_data, w_pars[1], w_pars[2], w_pars[3])
return(o_loglik + w_loglik)
}
# Initial estimates (method of moments for the Gumbel case)
sigma <- as.numeric(sqrt(6 * diag(var(owtemps))) / pi)
mu <- as.numeric(colMeans(owtemps) - 0.57722 * sigma)
init <- c(mean(mu), -diff(mu) / 2, mean(sigma), -diff(sigma) / 2, 0, 0)
# Log-likelihood adjustment of the full model
par_names <- c("mu[0]", "mu[1]", "sigma[0]", "sigma[1]", "xi[0]", "xi[1]")
large <- adjust_loglik(gev_loglik, data = owtemps, init = init,
par_names = par_names)
# 95% likelihood-based confidence intervals, vertically adjusted
large_v <- conf_intervals(large, which_pars = c("xi[0]", "xi[1]"))
#> Waiting for profiling to be done...
large_v
#> Model: gev_loglik
#>
#> 95% confidence intervals, adjusted loglikelihod with type = ''vertical''
#>
#> Symmetric:
#> lower upper
#> xi[0] -0.27623 -0.12166
#> xi[1] -0.15939 -0.01731
#>
#> Profile likelihood-based:
#> lower upper
#> xi[0] -0.27410 -0.11572
#> xi[1] -0.16519 -0.02002
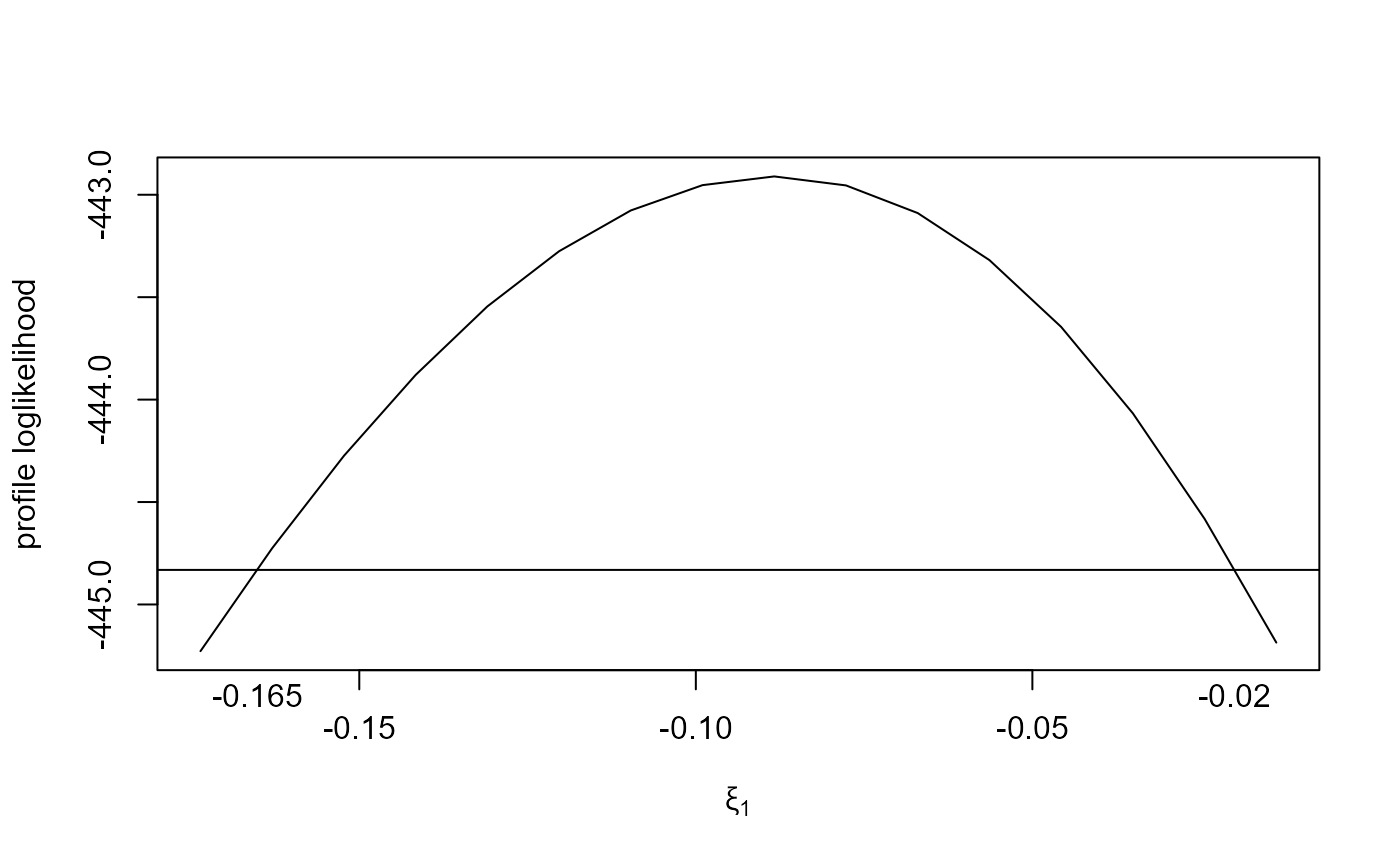
plot(large_v)
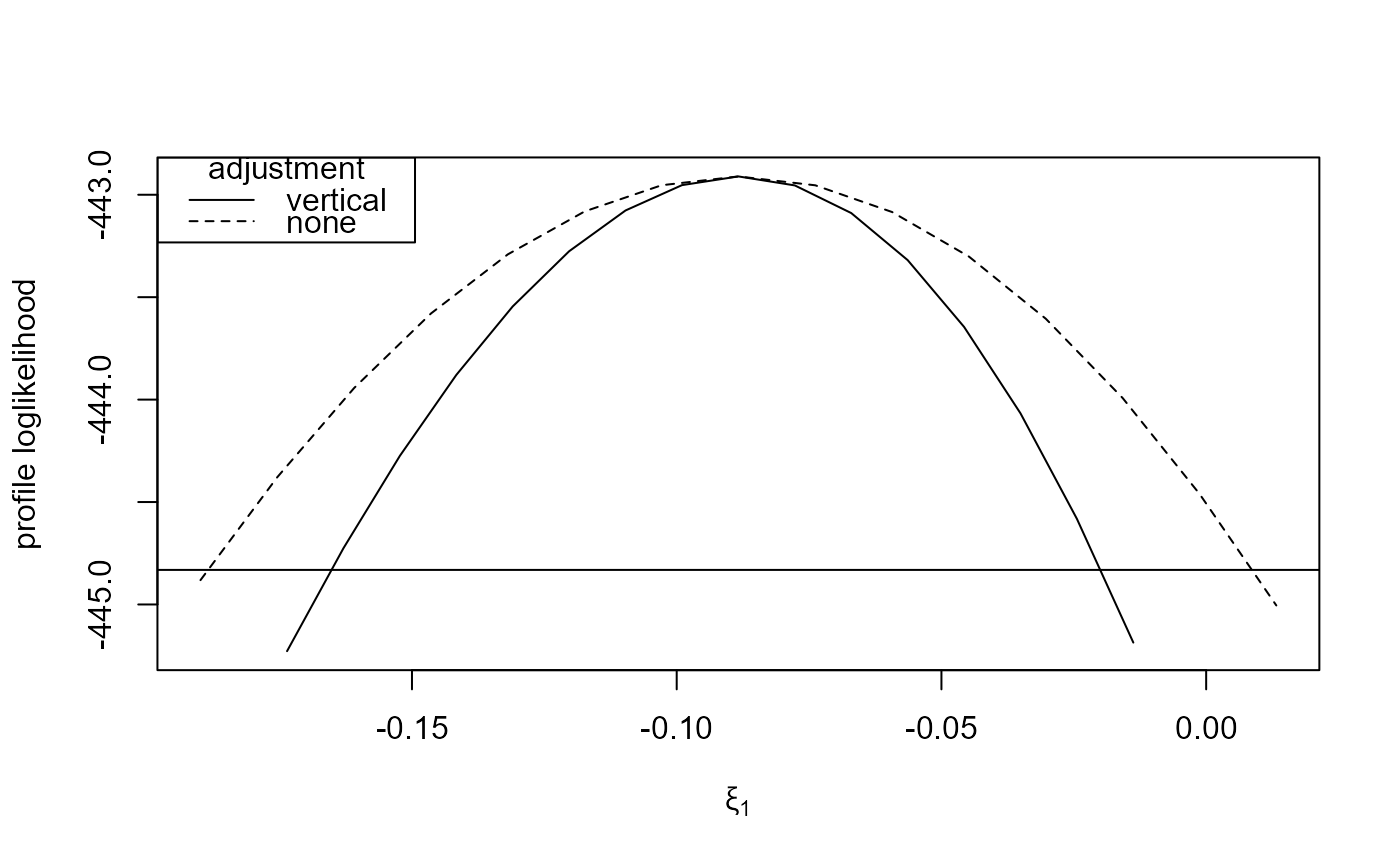
 plot(large_v, which_par = "xi[1]")
plot(large_v, which_par = "xi[1]")
 # \donttest{
# Unadjusted
large_none <- conf_intervals(large, which_pars = c("xi[0]", "xi[1]"),
type = "none")
#> Waiting for profiling to be done...
large_none
#> Model: gev_loglik
#>
#> 95% confidence intervals, independence loglikelihood
#>
#> Symmetric:
#> lower upper
#> xi[0] -0.295708 -0.102188
#> xi[1] -0.185107 0.008412
#>
#> Profile likelihood-based:
#> lower upper
#> xi[0] -0.289574 -0.094328
#> xi[1] -0.188502 0.008628
plot(large_v, large_none)
# \donttest{
# Unadjusted
large_none <- conf_intervals(large, which_pars = c("xi[0]", "xi[1]"),
type = "none")
#> Waiting for profiling to be done...
large_none
#> Model: gev_loglik
#>
#> 95% confidence intervals, independence loglikelihood
#>
#> Symmetric:
#> lower upper
#> xi[0] -0.295708 -0.102188
#> xi[1] -0.185107 0.008412
#>
#> Profile likelihood-based:
#> lower upper
#> xi[0] -0.289574 -0.094328
#> xi[1] -0.188502 0.008628
plot(large_v, large_none)
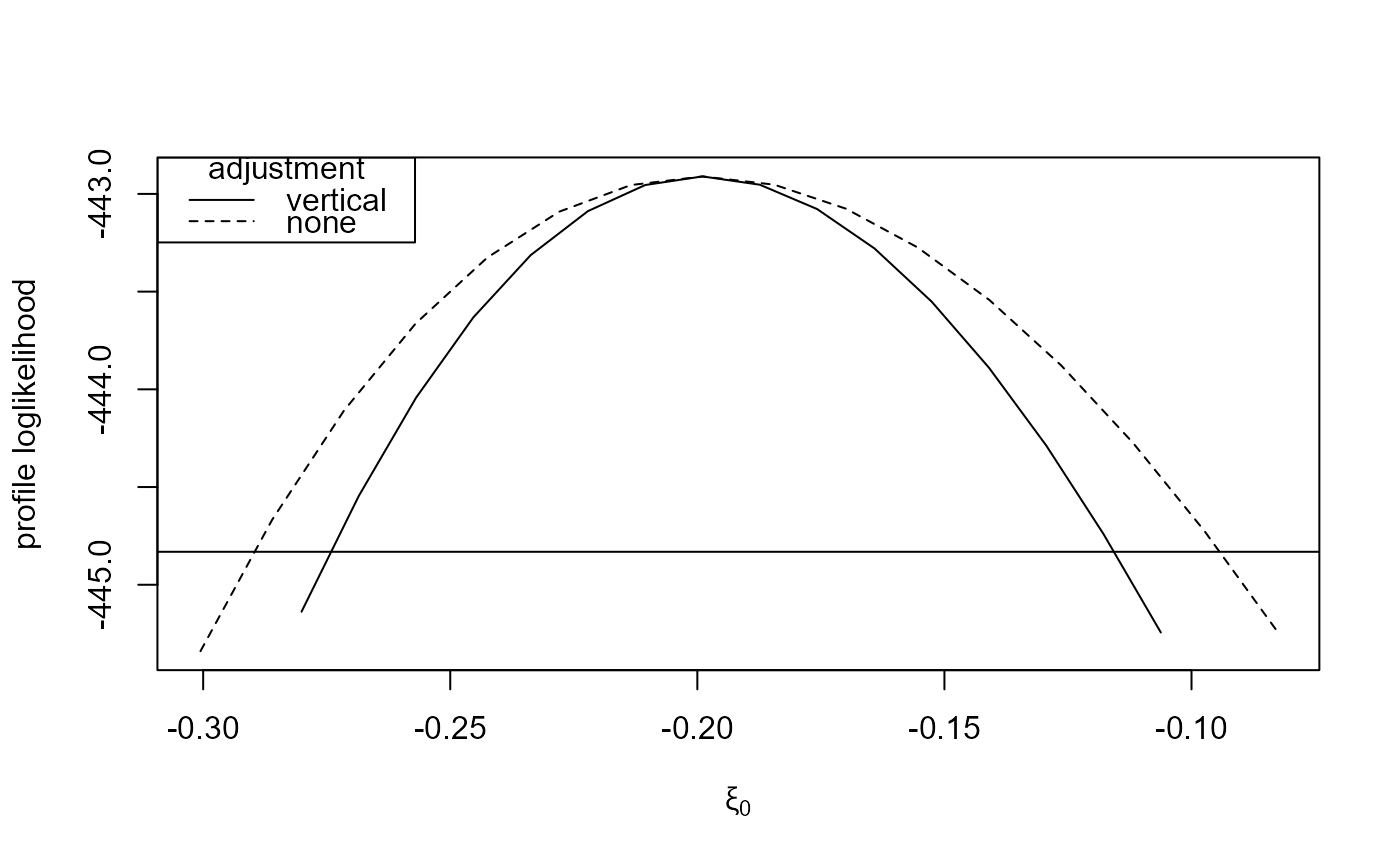 plot(large_v, large_none, which_par = "xi[1]")
plot(large_v, large_none, which_par = "xi[1]")
 # }
# --------- Misspecified Poisson model for negative binomial data ----------
# ... following Section 5.1 of the "Object-Oriented Computation of Sandwich
# Estimators" vignette of the sandwich package
# https://cran.r-project.org/web/packages/sandwich/vignettes/sandwich-OOP.pdf
# Simulate data
set.seed(123)
x <- rnorm(250)
y <- rnbinom(250, mu = exp(1 + x), size = 1)
# Fit misspecified Poisson model
fm_pois <- glm(y ~ x + I(x^2), family = poisson)
summary(fm_pois)$coefficients
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 1.06326821 0.04135723 25.709367 9.184267e-146
#> x 0.99607219 0.05353446 18.606186 2.862861e-77
#> I(x^2) -0.04912373 0.02314608 -2.122335 3.380961e-02
# Contributions to the independence loglikelihood
pois_glm_loglik <- function(pars, y, x) {
log_mu <- pars[1] + pars[2] * x + pars[3] * x ^ 2
return(dpois(y, lambda = exp(log_mu), log = TRUE))
}
pars <- c("alpha", "beta", "gamma")
pois_quad <- adjust_loglik(pois_glm_loglik, y = y, x = x, par_names = pars)
conf_intervals(pois_quad)
#> Waiting for profiling to be done...
#> Model: pois_glm_loglik
#>
#> 95% confidence intervals, adjusted loglikelihod with type = ''vertical''
#>
#> Symmetric:
#> lower upper
#> alpha 0.89907 1.22747
#> beta 0.78986 1.20231
#> gamma -0.12024 0.02199
#>
#> Profile likelihood-based:
#> lower upper
#> alpha 0.8954 1.2232
#> beta 0.7877 1.1991
#> gamma -0.1198 0.0222
# }
# --------- Misspecified Poisson model for negative binomial data ----------
# ... following Section 5.1 of the "Object-Oriented Computation of Sandwich
# Estimators" vignette of the sandwich package
# https://cran.r-project.org/web/packages/sandwich/vignettes/sandwich-OOP.pdf
# Simulate data
set.seed(123)
x <- rnorm(250)
y <- rnbinom(250, mu = exp(1 + x), size = 1)
# Fit misspecified Poisson model
fm_pois <- glm(y ~ x + I(x^2), family = poisson)
summary(fm_pois)$coefficients
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 1.06326821 0.04135723 25.709367 9.184267e-146
#> x 0.99607219 0.05353446 18.606186 2.862861e-77
#> I(x^2) -0.04912373 0.02314608 -2.122335 3.380961e-02
# Contributions to the independence loglikelihood
pois_glm_loglik <- function(pars, y, x) {
log_mu <- pars[1] + pars[2] * x + pars[3] * x ^ 2
return(dpois(y, lambda = exp(log_mu), log = TRUE))
}
pars <- c("alpha", "beta", "gamma")
pois_quad <- adjust_loglik(pois_glm_loglik, y = y, x = x, par_names = pars)
conf_intervals(pois_quad)
#> Waiting for profiling to be done...
#> Model: pois_glm_loglik
#>
#> 95% confidence intervals, adjusted loglikelihod with type = ''vertical''
#>
#> Symmetric:
#> lower upper
#> alpha 0.89907 1.22747
#> beta 0.78986 1.20231
#> gamma -0.12024 0.02199
#>
#> Profile likelihood-based:
#> lower upper
#> alpha 0.8954 1.2232
#> beta 0.7877 1.1991
#> gamma -0.1198 0.0222