Produces a diagnostic plot to assist in the selection of an extreme value threshold in the case where the data can be treated as independent and identically distributed (i.i.d.) observations. For example, it could be that these observations are the cluster maxima resulting from the declustering of time series data. The predictive ability of models fitted using each of a user-supplied set of thresholds is assessed using leave-one-out cross-validation in a Bayesian setup. These models are based on a Generalized Pareto (GP) distribution for threshold excesses and a binomial model for the probability of threshold exceedance. See Northrop et al. (2017) for details.
Arguments
- data
A numeric vector of observations. Any missing values will be removed. The argument
npy(see below) may be supplied as an attribute ofdatausingattr(data, "npy") <- value, wherevalueis the value ofnpy(seeattr). Ifnpyis supplied twice, as bothattr(data, "npy")) and using thenpyargument, then the former is used.- u_vec
A numeric vector. A vector of training thresholds at which inferences are made from a binomial-GP model. These could be set at sample quantiles of
datausingquantile. Any duplicated values will be removed.- ...
Further (optional) arguments to be passed to the
revdbayesfunctionrpost_rcpp(orrpost), which use the generalized ratio-of-uniforms method to simulate from extreme value posterior distributions. In particular:n. The size of the posterior sample used to perform predictive inference. Default:n = 1000.prior. A prior for GP parameters to be passed to the revdbayes functionset_prior. Can be either a character scalar that chooses an in-built prior, or a user-supplied R function or pointer to a compiled C++ function. See theset_priordocumentation for details of the in-built priors. See the revdbayes vignette Faster simulation using revdbayes for information about creating a pointer to a C++ function. See also the Examples section.If the user supplies an R function then
rpostwill be used for posterior simulation, rather than (the faster)rpost_rcpp, regardless of the input value ofuse_rcpp.Default:
prior = "mdi"witha = 0.6andmin_xi = -1. This particular prior is studied in Northrop et al. (2017).h_prior. A list of further arguments (hyperparameters) for the GP prior specified inprior.bin_prior. A prior for the threshold exceedance probability \(p\) to be passed to the revdbayes functionset_bin_prior. Can either be a character scalar that chooses an in-built prior, or a user_supplied R function.Default:
prior = "jeffreys", i.e. Beta(1/2, 1/2).h_bin_prior. A list of further arguments (hyperparameters) for the binomial prior specified inbin_prior. See theset_bin_priordocumentation for details of the in-built priors.trans. A character scalar: either"none"or"BC". Seerpost_rcppfor details. The default is"none", which is usually faster than"BC". However, if there are very few threshold excesses then usingtrans = "BC"can make the optimizations involved in the generalized ratio-of-uniforms algorithm more stable. If usingtrans = "none"produces an error for a particular posterior simulation thentrans = "BC"is used instead.
- n_v
A numeric scalar. Each of the
n_vlargest values inu_vecwill be used (separately) as a validation threshold for the training thresholds inu_vecthat lie at or below that validation threshold. Ifn_v = 1then all the training thresholds are used with validation performed using the thresholdu_vec[length(u_vec)]. Ifn_v = 2then, in addition, the assessment is performed usingu_vec[1], ..., u_vec[length(u_vec) - 1]with validation thresholdu_vec[length(u_vec) - 1], and so on.- npy
A numeric scalar. The mean number of observations per year of data, after excluding any missing values, i.e. the number of non-missing observations divided by total number of years of non-missing data. May be supplied using as an attribute
attr(data, "npy")ofdatainstead.The value of
npydoes not affect any calculation inithresh, it only affects subsequent extreme value inferences usingpredict.ithresh. However, settingnpyin the call torpost, or as an attribute ofdataavoids the need to supplynpywhen callingpredict.ithresh.- use_rcpp
A logical scalar. If
TRUE(the default) the revdbayes functionrpost_rcppis used for posterior simulation. IfFALSE, or if the user supplies an R function to set the prior for GP parameters, the (slower) functionrpostis used.
Value
An object (list) of class "ithresh", containing the
components
pred_perf: A numeric matrix withlength(u_vec)rows andn_vcolumns. Each column contains the values of the measure of predictive performance. Entries corresponding to cases where the training threshold is above the validation threshold will beNA.u_vec: The argumentu_vectoithresh.v_vec: A numeric vector. The validation thresholds implied by the argumentn_vtoithresh.u_ps: A numeric vector. The approximate levels of the sample quantiles to which the values inu_veccorrespond, i.e. the approximate percentage of the data the lie at or below each element inu_vec.v_ps: A numeric vector. The values inu_psthat correspond to the validation thresholds.sim_vals: A numeric matrix with 4 columns andnxlength(u_vec)rows. The \(j\)th block ofnrows contains in columns 1-3 the posterior samples of the threshold exceedance probability, the GP scale parameter and the GP shape parameter respectively, based on training thresholdu_vec[j], and in column 4 the value of \(j\).n: A numeric scalar. The value ofn.npy: A numeric scalar. The value ofnpy.data: The argumentdatatoithreshdetailed above, with any missing values removed.use_rcpp: A logical scalar indicating whetherrpost_rcpp(use_rcpp = TRUE) orrpost(use_rcpp = FALSE) was used for posterior simulation.for_post: A list containing arguments with whichrpost_rcpp(orrpost) was called, including any user-supplied arguments to these functions.call:The call toithresh.
Details
For a given threshold in u_vec:
the number of values in
datathat exceed the threshold, and the amounts (the threshold excesses) by which these value exceed the threshold are calculated;rpost_rcpp(orrpost) is used to sample from the posterior distributions of the parameters of a GP model for the threshold excesses and a binomial model for the probability of threshold exceedance;the ability of this binomial-GP model to predict data thresholded at the validation threshold(s) specified by
n_vis assessed using leave-one-out cross-validation (the measure of this is given in equation (7) of Northrop et al. (2017).
See Northrop et al. (2017) and the introductory threshr vignette for further details and examples.
References
Northrop, P.J. and Attalides, N. (2016) Posterior propriety in Bayesian extreme value analyses using reference priors Statistica Sinica, 26(2), 721–743 doi:10.5705/ss.2014.034 .
Northrop, P. J., Attalides, N. and Jonathan, P. (2017) Cross-validatory extreme value threshold selection and uncertainty with application to ocean storm severity. Journal of the Royal Statistical Society Series C: Applied Statistics, 66(1), 93-120. doi:10.1111/rssc.12159
Jonathan, P. and Ewans, K. (2013) Statistical modelling of extreme ocean environments for marine design : a review. Ocean Engineering, 62, 91-109. doi:10.1016/j.oceaneng.2013.01.004
See also
plot.ithresh for the S3 plot method for objects of
class ithresh.
summary.ithresh Summarizing measures of threshold
predictive performance.
predict.ithresh for predictive inference for the
largest value observed in N years.
rpost in the
revdbayes package for details of the arguments
that can be passed to
rpost_rcpp/rpost.
set_prior and
set_bin_prior in the
revdbayes package for details of how to set a
prior distributions for GP parameters and for the exceedance probability
\(p\).
Examples
# Note:
# 1. Smoother plots result from making n larger than the default n = 1000.
# 2. In some examples below validation thresholds rather higher than is
# advisable have been used, with far fewer excesses than the minimum of
# 50 suggested by Jonathan and Ewans (2013).
## North Sea significant wave heights, default prior -----------------------
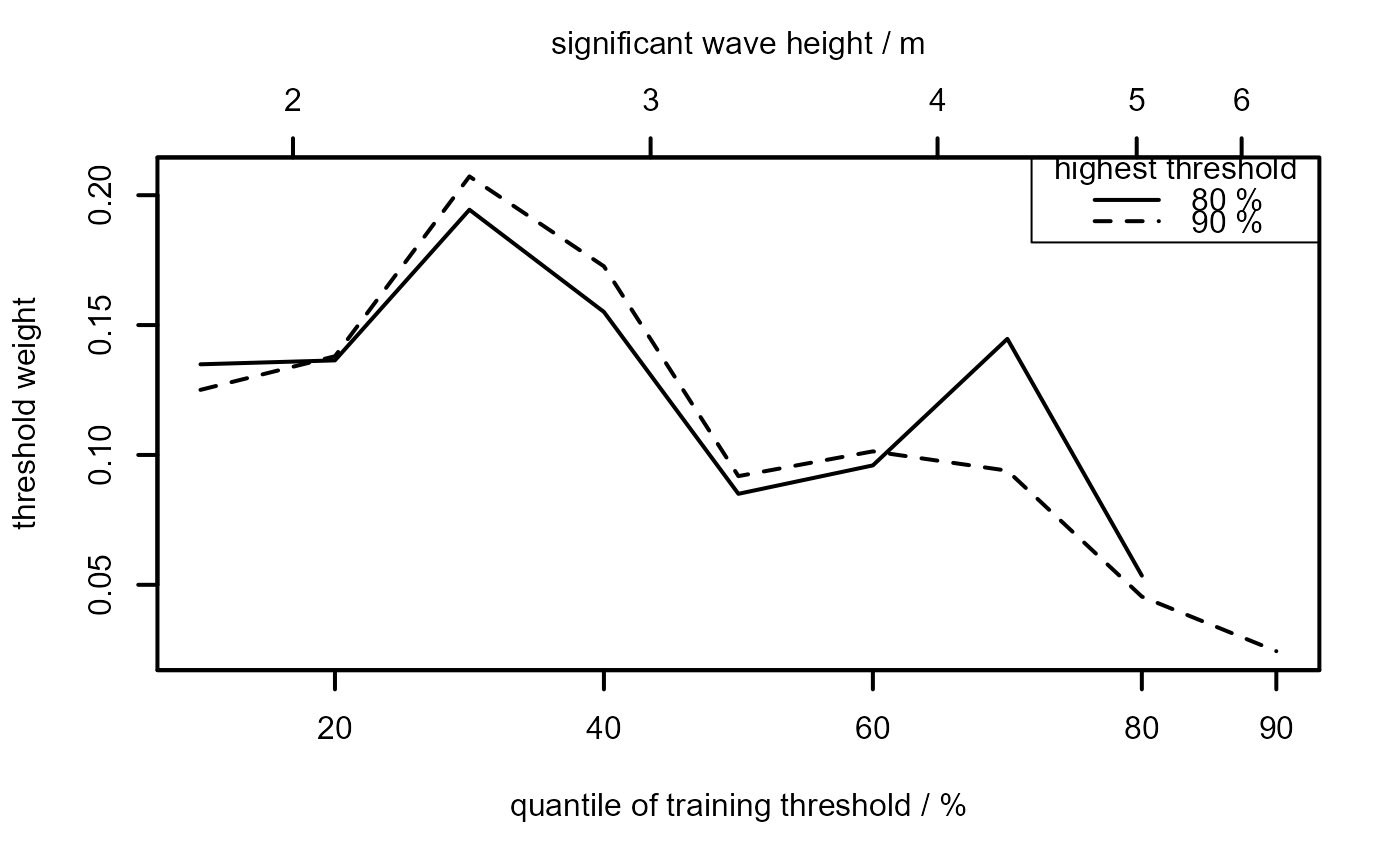
#' # A plot akin to the top left of Figure 7 in Northrop et al. (2017)
#' # ... but with fewer training thresholds
u_vec_ns <- quantile(ns, probs = seq(0.1, 0.9, by = 0.1))
ns_cv <- ithresh(data = ns, u_vec = u_vec_ns, n_v = 2)
plot(ns_cv, lwd = 2, add_legend = TRUE, legend_pos = "topright")
mtext("significant wave height / m", side = 3, line = 2.5)
 ## Gulf of Mexico significant wave heights, default prior ------------------
u_vec_gom <- quantile(gom, probs = seq(0.2, 0.9, by = 0.1))
# Setting a prior using its name and parameter value(s) --------------------
# This example gives the same prior as the default
gom_cv <- ithresh(data = gom, u_vec = u_vec_gom, n_v = 2, prior = "mdi",
h_prior = list(a = 0.6))
## Setting a user-defined (log-)prior R function ---------------------------
# This example also gives the same prior as the default
# (It will take longer to run than the example above because ithresh detects
# that the prior is an R function and sets use_rcpp to FALSE.)
# \donttest{
user_prior <- function(pars, a, min_xi = -1) {
if (pars[1] <= 0 | pars[2] < min_xi) {
return(-Inf)
}
return(-log(pars[1]) - a * pars[2])
}
user_bin_prior <- function(p, ab) {
return(stats::dbeta(p, shape1 = ab[1], shape2 = ab[2], log = TRUE))
}
gom_cv <- ithresh(data = gom, u_vec = u_vec_gom, n_v = 2, prior = user_prior,
h_prior = list(a = 0.6), bin_prior = user_bin_prior,
h_bin_prior = list(ab = c(1 / 2, 1 / 2)))
# }
## Setting a user-defined (log-)prior (pointer to a) C++ function ----------
# We make use of a C++ function and function create_prior_xptr() to create
# the required pointer from the revdbayes package
prior_ptr <- revdbayes::create_prior_xptr("gp_flat")
gom_cv <- ithresh(data = gom, u_vec = u_vec_gom, n_v = 2, prior = prior_ptr,
h_prior = list(min_xi = -1))
## Gulf of Mexico significant wave heights, default prior ------------------
u_vec_gom <- quantile(gom, probs = seq(0.2, 0.9, by = 0.1))
# Setting a prior using its name and parameter value(s) --------------------
# This example gives the same prior as the default
gom_cv <- ithresh(data = gom, u_vec = u_vec_gom, n_v = 2, prior = "mdi",
h_prior = list(a = 0.6))
## Setting a user-defined (log-)prior R function ---------------------------
# This example also gives the same prior as the default
# (It will take longer to run than the example above because ithresh detects
# that the prior is an R function and sets use_rcpp to FALSE.)
# \donttest{
user_prior <- function(pars, a, min_xi = -1) {
if (pars[1] <= 0 | pars[2] < min_xi) {
return(-Inf)
}
return(-log(pars[1]) - a * pars[2])
}
user_bin_prior <- function(p, ab) {
return(stats::dbeta(p, shape1 = ab[1], shape2 = ab[2], log = TRUE))
}
gom_cv <- ithresh(data = gom, u_vec = u_vec_gom, n_v = 2, prior = user_prior,
h_prior = list(a = 0.6), bin_prior = user_bin_prior,
h_bin_prior = list(ab = c(1 / 2, 1 / 2)))
# }
## Setting a user-defined (log-)prior (pointer to a) C++ function ----------
# We make use of a C++ function and function create_prior_xptr() to create
# the required pointer from the revdbayes package
prior_ptr <- revdbayes::create_prior_xptr("gp_flat")
gom_cv <- ithresh(data = gom, u_vec = u_vec_gom, n_v = 2, prior = prior_ptr,
h_prior = list(min_xi = -1))